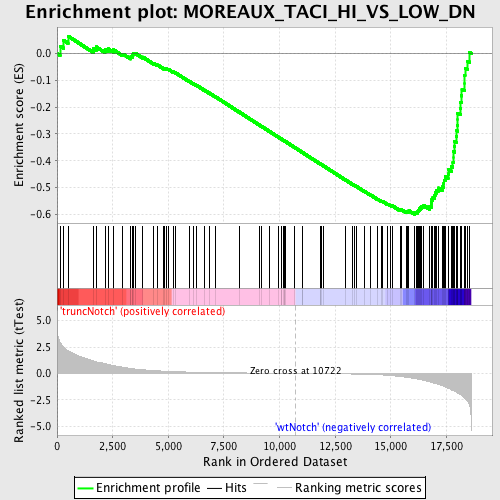
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | MOREAUX_TACI_HI_VS_LOW_DN |
| Enrichment Score (ES) | -0.6008503 |
| Normalized Enrichment Score (NES) | -1.7976893 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.044448193 |
| FWER p-Value | 0.202 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPL43 | 360014 5360673 | 133 | 2.914 | 0.0284 | No | ||
| 2 | TYMS | 940450 1940068 3710008 5570546 | 288 | 2.473 | 0.0503 | No | ||
| 3 | ERCC1 | 3800010 4590132 4670397 | 493 | 2.149 | 0.0656 | No | ||
| 4 | RUVBL2 | 1190377 | 1632 | 1.182 | 0.0185 | No | ||
| 5 | FIBP | 2510450 | 1755 | 1.103 | 0.0254 | No | ||
| 6 | JTV1 | 460324 | 2153 | 0.921 | 0.0152 | No | ||
| 7 | TIMM50 | 1500139 | 2297 | 0.857 | 0.0179 | No | ||
| 8 | GPR89A | 780315 1580368 | 2527 | 0.754 | 0.0148 | No | ||
| 9 | MRPS12 | 4210707 6110341 | 2958 | 0.579 | -0.0014 | No | ||
| 10 | NDUFA13 | 6220021 | 3303 | 0.462 | -0.0143 | No | ||
| 11 | MCM2 | 5050139 | 3319 | 0.459 | -0.0095 | No | ||
| 12 | BUB3 | 3170546 | 3396 | 0.440 | -0.0082 | No | ||
| 13 | RACGAP1 | 3990162 6620736 | 3408 | 0.437 | -0.0035 | No | ||
| 14 | LSM4 | 3130180 | 3437 | 0.430 | 0.0002 | No | ||
| 15 | PAPOLA | 4200286 4850112 | 3506 | 0.415 | 0.0016 | No | ||
| 16 | MATR3 | 1940170 5340278 | 3846 | 0.346 | -0.0124 | No | ||
| 17 | SPAST | 460603 4070471 4210132 4670338 | 4351 | 0.266 | -0.0364 | No | ||
| 18 | EIF3S4 | 3840017 6400519 | 4512 | 0.242 | -0.0421 | No | ||
| 19 | PPP2R5C | 3310121 6650672 | 4798 | 0.212 | -0.0549 | No | ||
| 20 | NNT | 540253 1170471 5550092 6760397 | 4822 | 0.209 | -0.0536 | No | ||
| 21 | PRKDC | 1400072 4230541 | 4927 | 0.199 | -0.0568 | No | ||
| 22 | EBNA1BP2 | 50369 1400433 6040019 | 4987 | 0.193 | -0.0576 | No | ||
| 23 | TTRAP | 6400722 | 5218 | 0.172 | -0.0680 | No | ||
| 24 | CYP2R1 | 2360010 | 5302 | 0.165 | -0.0704 | No | ||
| 25 | MYLIP | 50717 4050672 | 5932 | 0.123 | -0.1029 | No | ||
| 26 | SLC33A1 | 2100711 | 6151 | 0.111 | -0.1133 | No | ||
| 27 | TRIB1 | 2320435 | 6277 | 0.106 | -0.1188 | No | ||
| 28 | SLC16A1 | 50433 | 6612 | 0.091 | -0.1357 | No | ||
| 29 | LAP3 | 2570064 | 6836 | 0.082 | -0.1468 | No | ||
| 30 | MET | 2690348 | 7105 | 0.073 | -0.1604 | No | ||
| 31 | PSMA3 | 5900047 7040161 | 8176 | 0.045 | -0.2176 | No | ||
| 32 | THYN1 | 2940632 | 9099 | 0.027 | -0.2671 | No | ||
| 33 | PAK1IP1 | 2230438 | 9170 | 0.026 | -0.2706 | No | ||
| 34 | IFT74 | 6980170 | 9548 | 0.019 | -0.2907 | No | ||
| 35 | LSM5 | 4540398 | 9935 | 0.013 | -0.3114 | No | ||
| 36 | MCM3AP | 4280465 | 10073 | 0.011 | -0.3187 | No | ||
| 37 | DARS | 580427 3840139 | 10103 | 0.010 | -0.3201 | No | ||
| 38 | IFT57 | 1190541 2360292 6400280 | 10196 | 0.008 | -0.3250 | No | ||
| 39 | CCDC5 | 2480040 | 10229 | 0.008 | -0.3266 | No | ||
| 40 | TMEM165 | 1230014 | 10232 | 0.008 | -0.3266 | No | ||
| 41 | STAM2 | 730731 2120377 | 10259 | 0.007 | -0.3279 | No | ||
| 42 | TERF1 | 2340113 | 10676 | 0.001 | -0.3504 | No | ||
| 43 | PSMD8 | 630142 | 11013 | -0.005 | -0.3685 | No | ||
| 44 | ABHD3 | 2370091 | 11016 | -0.005 | -0.3685 | No | ||
| 45 | RAN | 2260446 4590647 | 11827 | -0.019 | -0.4121 | No | ||
| 46 | ANAPC10 | 870086 1170037 2260129 | 11843 | -0.019 | -0.4126 | No | ||
| 47 | DHX29 | 6180300 | 11870 | -0.020 | -0.4138 | No | ||
| 48 | PDCD6 | 4760239 | 11977 | -0.022 | -0.4193 | No | ||
| 49 | GEMIN5 | 3120577 | 12968 | -0.047 | -0.4722 | No | ||
| 50 | CHEK1 | 50167 1940300 | 13289 | -0.059 | -0.4887 | No | ||
| 51 | TATDN1 | 2810722 | 13377 | -0.062 | -0.4927 | No | ||
| 52 | CDC23 | 3190593 | 13459 | -0.066 | -0.4963 | No | ||
| 53 | PAPD4 | 1580673 730605 5270112 | 13811 | -0.083 | -0.5142 | No | ||
| 54 | ACTR6 | 4570079 | 14101 | -0.101 | -0.5286 | No | ||
| 55 | MINA | 6660152 5290181 | 14398 | -0.126 | -0.5430 | No | ||
| 56 | CCT2 | 1690113 | 14576 | -0.142 | -0.5509 | No | ||
| 57 | EZH2 | 6130605 6380524 | 14633 | -0.150 | -0.5521 | No | ||
| 58 | TLE4 | 1450064 1500519 | 14863 | -0.181 | -0.5622 | No | ||
| 59 | NUDT5 | 6940408 | 14990 | -0.201 | -0.5666 | No | ||
| 60 | AASDHPPT | 6200671 | 15064 | -0.213 | -0.5679 | No | ||
| 61 | IARS | 460528 1580739 1690008 | 15415 | -0.287 | -0.5833 | No | ||
| 62 | RNF7 | 2030242 | 15465 | -0.298 | -0.5823 | No | ||
| 63 | COMMD4 | 510110 | 15700 | -0.368 | -0.5905 | No | ||
| 64 | RNF111 | 2970072 3140112 5130647 | 15769 | -0.389 | -0.5894 | No | ||
| 65 | ANKRD17 | 2120402 3850035 6400044 | 15814 | -0.408 | -0.5868 | No | ||
| 66 | MINPP1 | 870301 2230731 2940148 5860736 6110112 | 16075 | -0.491 | -0.5948 | Yes | ||
| 67 | PSMC4 | 580050 1580025 | 16153 | -0.518 | -0.5927 | Yes | ||
| 68 | PPP1R12A | 4070066 4590162 | 16212 | -0.543 | -0.5892 | Yes | ||
| 69 | EFTUD2 | 2470707 4610148 | 16247 | -0.554 | -0.5842 | Yes | ||
| 70 | BARD1 | 3170348 | 16293 | -0.574 | -0.5797 | Yes | ||
| 71 | PCID1 | 6020112 | 16334 | -0.588 | -0.5746 | Yes | ||
| 72 | MRPL19 | 6350400 7000121 | 16385 | -0.608 | -0.5699 | Yes | ||
| 73 | SNRPD1 | 4480162 | 16482 | -0.653 | -0.5671 | Yes | ||
| 74 | KEAP1 | 1570121 2900546 3840576 | 16747 | -0.805 | -0.5715 | Yes | ||
| 75 | KRIT1 | 2640500 4230435 4730184 | 16823 | -0.851 | -0.5652 | Yes | ||
| 76 | CCT5 | 1050670 | 16829 | -0.853 | -0.5550 | Yes | ||
| 77 | SHFM1 | 1570114 | 16843 | -0.860 | -0.5452 | Yes | ||
| 78 | CIRH1A | 2340372 | 16891 | -0.891 | -0.5369 | Yes | ||
| 79 | ASF1A | 7050020 | 16955 | -0.931 | -0.5289 | Yes | ||
| 80 | SPATA5L1 | 6290692 | 16991 | -0.961 | -0.5191 | Yes | ||
| 81 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 17037 | -0.982 | -0.5095 | Yes | ||
| 82 | SMARCA4 | 6980528 | 17126 | -1.018 | -0.5018 | Yes | ||
| 83 | SF3B2 | 6590576 | 17310 | -1.148 | -0.4977 | Yes | ||
| 84 | PFDN4 | 4010286 | 17379 | -1.214 | -0.4865 | Yes | ||
| 85 | CHCHD1 | 110706 | 17390 | -1.222 | -0.4721 | Yes | ||
| 86 | C1QBP | 3170452 | 17457 | -1.274 | -0.4601 | Yes | ||
| 87 | UPF3A | 2360333 5340008 | 17574 | -1.390 | -0.4494 | Yes | ||
| 88 | ORC5L | 1940133 1940711 | 17603 | -1.414 | -0.4337 | Yes | ||
| 89 | KLF13 | 4670563 | 17724 | -1.552 | -0.4212 | Yes | ||
| 90 | PAIP1 | 3870576 | 17791 | -1.625 | -0.4049 | Yes | ||
| 91 | ILF3 | 940722 3190647 6520110 | 17823 | -1.657 | -0.3863 | Yes | ||
| 92 | HNRPAB | 540504 | 17825 | -1.660 | -0.3661 | Yes | ||
| 93 | SGOL2 | 2030338 | 17861 | -1.693 | -0.3473 | Yes | ||
| 94 | BTBD1 | 5720619 | 17872 | -1.705 | -0.3270 | Yes | ||
| 95 | IREB2 | 1570202 2640114 | 17939 | -1.775 | -0.3089 | Yes | ||
| 96 | FANCL | 2060520 5220066 5270164 | 17965 | -1.793 | -0.2883 | Yes | ||
| 97 | FEM1B | 1230110 | 17994 | -1.837 | -0.2674 | Yes | ||
| 98 | RSF1 | 1580097 | 18005 | -1.851 | -0.2453 | Yes | ||
| 99 | ANKRD49 | 780102 | 18008 | -1.853 | -0.2228 | Yes | ||
| 100 | DDX31 | 430279 6450576 | 18124 | -2.001 | -0.2046 | Yes | ||
| 101 | IK | 1690039 | 18140 | -2.024 | -0.1806 | Yes | ||
| 102 | DDX1 | 3450047 | 18181 | -2.082 | -0.1574 | Yes | ||
| 103 | MRPL50 | 1740465 | 18198 | -2.107 | -0.1325 | Yes | ||
| 104 | NASP | 2260139 2940369 5130707 | 18318 | -2.326 | -0.1105 | Yes | ||
| 105 | TSHZ1 | 4780427 | 18321 | -2.329 | -0.0822 | Yes | ||
| 106 | NMT1 | 5810152 | 18336 | -2.373 | -0.0539 | Yes | ||
| 107 | SNX19 | 4200433 4230609 | 18458 | -2.660 | -0.0280 | Yes | ||
| 108 | DDX18 | 6980619 | 18535 | -2.983 | 0.0044 | Yes |

