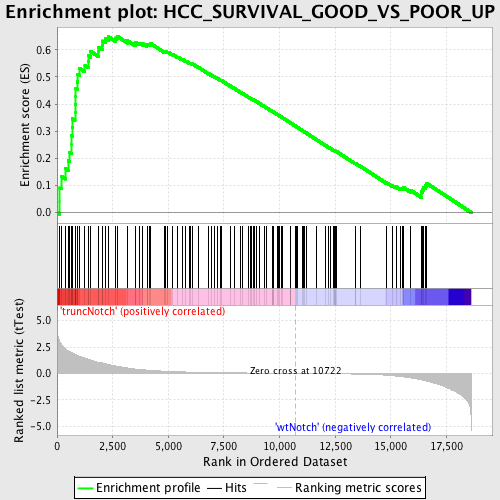
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | truncNotch |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_UP |
| Enrichment Score (ES) | 0.6504661 |
| Normalized Enrichment Score (NES) | 1.8316383 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.013921606 |
| FWER p-Value | 0.016 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | C2 | 5390465 | 126 | 2.930 | 0.0420 | Yes | ||
| 2 | ACADS | 110746 | 127 | 2.930 | 0.0908 | Yes | ||
| 3 | DCXR | 4730551 | 185 | 2.757 | 0.1337 | Yes | ||
| 4 | KHK | 1090204 3870204 | 370 | 2.330 | 0.1626 | Yes | ||
| 5 | EGLN2 | 540086 | 497 | 2.139 | 0.1914 | Yes | ||
| 6 | PCCB | 50670 | 565 | 2.056 | 0.2221 | Yes | ||
| 7 | EPHX1 | 1740136 | 640 | 1.976 | 0.2510 | Yes | ||
| 8 | ABHD6 | 630717 | 643 | 1.974 | 0.2838 | Yes | ||
| 9 | CRYL1 | 1340427 | 675 | 1.935 | 0.3143 | Yes | ||
| 10 | SLC25A10 | 1400609 | 691 | 1.919 | 0.3455 | Yes | ||
| 11 | CPT2 | 3940504 6180075 | 805 | 1.808 | 0.3695 | Yes | ||
| 12 | MAP4K1 | 4060692 | 810 | 1.804 | 0.3994 | Yes | ||
| 13 | NDUFS2 | 4850020 6200402 | 837 | 1.771 | 0.4275 | Yes | ||
| 14 | SERPING1 | 5550440 | 842 | 1.761 | 0.4566 | Yes | ||
| 15 | ALDH4A1 | 2450450 | 902 | 1.712 | 0.4819 | Yes | ||
| 16 | PINK1 | 380044 580577 | 930 | 1.691 | 0.5087 | Yes | ||
| 17 | SERPINF1 | 7040367 | 994 | 1.624 | 0.5323 | Yes | ||
| 18 | ENPP5 | 1780338 | 1251 | 1.435 | 0.5424 | Yes | ||
| 19 | NDRG2 | 450403 | 1396 | 1.340 | 0.5570 | Yes | ||
| 20 | ECHS1 | 2970184 | 1413 | 1.330 | 0.5783 | Yes | ||
| 21 | SLC35D1 | 1740520 | 1509 | 1.261 | 0.5941 | Yes | ||
| 22 | C1QTNF4 | 60619 | 1864 | 1.045 | 0.5924 | Yes | ||
| 23 | SELENBP1 | 3850575 | 1869 | 1.041 | 0.6096 | Yes | ||
| 24 | ACOX2 | 450017 | 2040 | 0.977 | 0.6166 | Yes | ||
| 25 | PCYT2 | 2630092 4590278 | 2048 | 0.974 | 0.6325 | Yes | ||
| 26 | MVK | 1450717 | 2171 | 0.913 | 0.6411 | Yes | ||
| 27 | MASP2 | 360110 | 2313 | 0.848 | 0.6476 | Yes | ||
| 28 | G6PC | 430093 | 2610 | 0.716 | 0.6435 | Yes | ||
| 29 | IVD | 2510673 | 2692 | 0.678 | 0.6505 | Yes | ||
| 30 | PPAP2A | 1170397 3440288 3930270 | 3167 | 0.503 | 0.6332 | No | ||
| 31 | INSR | 1190504 | 3522 | 0.412 | 0.6210 | No | ||
| 32 | HAGH | 540358 | 3532 | 0.410 | 0.6273 | No | ||
| 33 | F12 | 1090215 3520736 | 3706 | 0.372 | 0.6242 | No | ||
| 34 | SLC38A3 | 5340142 | 3843 | 0.346 | 0.6226 | No | ||
| 35 | CRAT | 540020 2060364 3520148 | 4040 | 0.314 | 0.6172 | No | ||
| 36 | GLYAT | 6200239 | 4068 | 0.308 | 0.6209 | No | ||
| 37 | INSIG1 | 1500332 3450458 4540301 | 4153 | 0.292 | 0.6212 | No | ||
| 38 | AMT | 3180450 | 4216 | 0.282 | 0.6226 | No | ||
| 39 | RDH5 | 2970736 3940632 5340730 6370093 4610059 | 4833 | 0.209 | 0.5928 | No | ||
| 40 | VPREB1 | 3520280 | 4891 | 0.202 | 0.5931 | No | ||
| 41 | CPN2 | 6590291 | 4975 | 0.194 | 0.5918 | No | ||
| 42 | MSRA | 4570411 | 5182 | 0.175 | 0.5836 | No | ||
| 43 | GJB1 | 1240441 | 5422 | 0.156 | 0.5733 | No | ||
| 44 | PRDX6 | 4920397 6380601 | 5616 | 0.142 | 0.5652 | No | ||
| 45 | SERPIND1 | 6110059 | 5784 | 0.132 | 0.5584 | No | ||
| 46 | ABCG8 | 4780605 | 5969 | 0.121 | 0.5504 | No | ||
| 47 | SGPL1 | 4480059 | 6005 | 0.119 | 0.5505 | No | ||
| 48 | ABCC2 | 380202 5390300 | 6084 | 0.115 | 0.5482 | No | ||
| 49 | RGN | 6200095 | 6341 | 0.102 | 0.5361 | No | ||
| 50 | ZP3 | 4060767 | 6815 | 0.083 | 0.5119 | No | ||
| 51 | CPB2 | 4150167 6100333 6590735 | 6924 | 0.079 | 0.5074 | No | ||
| 52 | AQP9 | 2650047 | 7058 | 0.075 | 0.5014 | No | ||
| 53 | ABCA6 | 130538 1050706 | 7198 | 0.070 | 0.4951 | No | ||
| 54 | KLK3 | 1500278 | 7344 | 0.066 | 0.4884 | No | ||
| 55 | MUCDHL | 5720056 | 7398 | 0.064 | 0.4866 | No | ||
| 56 | F13B | 6220278 | 7777 | 0.055 | 0.4671 | No | ||
| 57 | DPYS | 3130670 | 7975 | 0.050 | 0.4573 | No | ||
| 58 | OTC | 1740494 | 8254 | 0.043 | 0.4430 | No | ||
| 59 | HAO1 | 7050093 | 8316 | 0.042 | 0.4404 | No | ||
| 60 | MPDZ | 130026 1990064 4050577 | 8596 | 0.037 | 0.4259 | No | ||
| 61 | LPIN1 | 1770358 3120059 | 8673 | 0.035 | 0.4224 | No | ||
| 62 | AMACR | 780215 | 8724 | 0.034 | 0.4202 | No | ||
| 63 | CDO1 | 2480279 | 8826 | 0.032 | 0.4153 | No | ||
| 64 | UGT1A6 | 4610368 6900086 4540092 2630075 | 8874 | 0.031 | 0.4133 | No | ||
| 65 | APCS | 2060500 | 8966 | 0.029 | 0.4088 | No | ||
| 66 | SERPINC1 | 1780717 | 9110 | 0.027 | 0.4016 | No | ||
| 67 | APOC3 | 2030168 | 9301 | 0.024 | 0.3917 | No | ||
| 68 | CES2 | 5340609 | 9390 | 0.022 | 0.3873 | No | ||
| 69 | SLC6A12 | 3170685 | 9664 | 0.018 | 0.3728 | No | ||
| 70 | RHBG | 2810736 | 9675 | 0.017 | 0.3726 | No | ||
| 71 | NF1 | 6980433 | 9703 | 0.017 | 0.3714 | No | ||
| 72 | AOX1 | 110082 6290450 | 9714 | 0.016 | 0.3711 | No | ||
| 73 | FMO3 | 2480369 | 9887 | 0.014 | 0.3621 | No | ||
| 74 | TTPA | 630576 | 9951 | 0.013 | 0.3589 | No | ||
| 75 | ANXA9 | 1190195 | 9995 | 0.012 | 0.3568 | No | ||
| 76 | PKLR | 1170400 2470114 | 10067 | 0.011 | 0.3531 | No | ||
| 77 | TTBK1 | 60605 1850722 | 10136 | 0.009 | 0.3496 | No | ||
| 78 | ITIH1 | 670044 | 10488 | 0.004 | 0.3307 | No | ||
| 79 | CUTL2 | 2940692 6220687 | 10490 | 0.004 | 0.3307 | No | ||
| 80 | PCK1 | 7000358 | 10495 | 0.004 | 0.3305 | No | ||
| 81 | APOA5 | 6130471 | 10717 | 0.000 | 0.3186 | No | ||
| 82 | PAH | 3800309 | 10759 | -0.001 | 0.3164 | No | ||
| 83 | PIPOX | 70750 | 10810 | -0.002 | 0.3137 | No | ||
| 84 | ABCG5 | 2810670 | 11034 | -0.005 | 0.3017 | No | ||
| 85 | SULT2A1 | 2650286 | 11067 | -0.006 | 0.3001 | No | ||
| 86 | BAAT | 730739 | 11098 | -0.006 | 0.2986 | No | ||
| 87 | MST1 | 1400403 | 11192 | -0.008 | 0.2937 | No | ||
| 88 | SALL1 | 5420020 7050195 | 11661 | -0.016 | 0.2687 | No | ||
| 89 | HYAL1 | 3850341 | 12050 | -0.024 | 0.2481 | No | ||
| 90 | CABIN1 | 6900446 | 12177 | -0.027 | 0.2417 | No | ||
| 91 | HPD | 3120519 | 12293 | -0.029 | 0.2360 | No | ||
| 92 | CES1 | 1400494 | 12418 | -0.032 | 0.2298 | No | ||
| 93 | SLC2A2 | 5130537 5720722 6770079 | 12450 | -0.033 | 0.2287 | No | ||
| 94 | MYRIP | 1580471 | 12483 | -0.033 | 0.2275 | No | ||
| 95 | CES3 | 5050167 3290458 | 12501 | -0.034 | 0.2272 | No | ||
| 96 | ASGR1 | 1850088 | 12525 | -0.034 | 0.2265 | No | ||
| 97 | MTSS1 | 780435 2370114 | 12568 | -0.035 | 0.2248 | No | ||
| 98 | DIO1 | 4570279 5290112 5420148 | 13390 | -0.063 | 0.1815 | No | ||
| 99 | HGD | 460390 | 13411 | -0.064 | 0.1815 | No | ||
| 100 | WNT11 | 1230278 | 13658 | -0.075 | 0.1694 | No | ||
| 101 | SLC27A5 | 2850717 | 14813 | -0.173 | 0.1100 | No | ||
| 102 | F10 | 3830450 | 15052 | -0.209 | 0.1006 | No | ||
| 103 | EHHADH | 6200315 | 15236 | -0.250 | 0.0949 | No | ||
| 104 | SPINK2 | 3990093 | 15414 | -0.287 | 0.0901 | No | ||
| 105 | IL6R | 520706 3800215 | 15534 | -0.318 | 0.0890 | No | ||
| 106 | AR | 6380167 | 15557 | -0.323 | 0.0931 | No | ||
| 107 | ITPR2 | 5360128 | 15879 | -0.428 | 0.0829 | No | ||
| 108 | ALAS1 | 6400440 | 16367 | -0.602 | 0.0666 | No | ||
| 109 | MRPL46 | 360736 | 16368 | -0.602 | 0.0767 | No | ||
| 110 | STARD10 | 1400619 4570170 | 16433 | -0.632 | 0.0837 | No | ||
| 111 | SEC14L2 | 2640370 | 16453 | -0.638 | 0.0933 | No | ||
| 112 | AMFR | 2810041 | 16555 | -0.687 | 0.0993 | No | ||
| 113 | SRD5A1 | 5910347 | 16605 | -0.720 | 0.1087 | No |