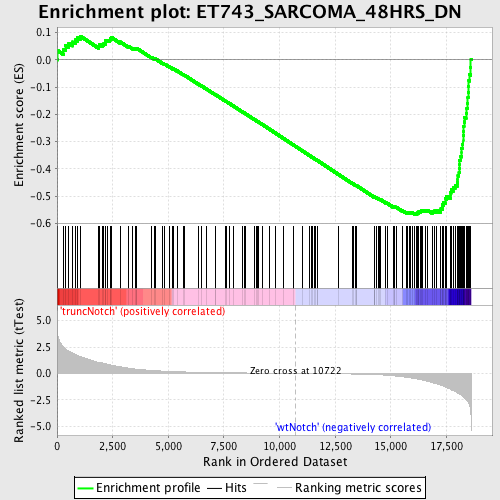
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ET743_SARCOMA_48HRS_DN |
| Enrichment Score (ES) | -0.5673441 |
| Normalized Enrichment Score (NES) | -1.7431602 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.044970233 |
| FWER p-Value | 0.446 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WNT5B | 5080162 | 10 | 4.536 | 0.0347 | No | ||
| 2 | KIAA0859 | 510497 770722 1340687 6840711 | 296 | 2.453 | 0.0383 | No | ||
| 3 | LHFPL2 | 450020 | 371 | 2.327 | 0.0524 | No | ||
| 4 | HSPE1 | 3170438 | 526 | 2.106 | 0.0605 | No | ||
| 5 | SLC2A1 | 2100609 | 705 | 1.907 | 0.0656 | No | ||
| 6 | RBM14 | 5340731 5690315 | 822 | 1.791 | 0.0733 | No | ||
| 7 | BAG2 | 2900324 | 925 | 1.693 | 0.0809 | No | ||
| 8 | SNIP1 | 5360092 | 1039 | 1.585 | 0.0871 | No | ||
| 9 | PANX1 | 6550632 | 1861 | 1.045 | 0.0508 | No | ||
| 10 | SIRPA | 130465 5700332 | 1898 | 1.028 | 0.0569 | No | ||
| 11 | TOP1 | 770471 4060632 6650324 | 2058 | 0.968 | 0.0558 | No | ||
| 12 | SON | 3120148 5860239 7040403 | 2102 | 0.945 | 0.0608 | No | ||
| 13 | TARDBP | 870390 2350093 | 2164 | 0.917 | 0.0646 | No | ||
| 14 | MID1IP1 | 2570739 | 2176 | 0.910 | 0.0711 | No | ||
| 15 | USP3 | 2060332 | 2271 | 0.870 | 0.0728 | No | ||
| 16 | DNAJA2 | 2630253 6860358 | 2377 | 0.817 | 0.0734 | No | ||
| 17 | ARFGAP1 | 4780364 5390358 | 2393 | 0.808 | 0.0789 | No | ||
| 18 | ZDHHC16 | 4230390 | 2446 | 0.783 | 0.0822 | No | ||
| 19 | DLG5 | 450215 | 2839 | 0.620 | 0.0658 | No | ||
| 20 | BLZF1 | 630088 | 3211 | 0.491 | 0.0495 | No | ||
| 21 | BUB3 | 3170546 | 3396 | 0.440 | 0.0430 | No | ||
| 22 | PDGFB | 3060440 6370008 | 3509 | 0.414 | 0.0401 | No | ||
| 23 | CLCN7 | 2970095 | 3551 | 0.407 | 0.0411 | No | ||
| 24 | EIF3S10 | 1190079 3840176 | 3569 | 0.403 | 0.0433 | No | ||
| 25 | MRPL1 | 430626 3130129 5220152 | 4223 | 0.280 | 0.0101 | No | ||
| 26 | ADSL | 5570484 | 4387 | 0.261 | 0.0033 | No | ||
| 27 | PSMA5 | 5390537 | 4404 | 0.258 | 0.0044 | No | ||
| 28 | THRAP3 | 6180309 | 4753 | 0.216 | -0.0127 | No | ||
| 29 | HSPA8 | 2120315 1050132 6550746 | 4841 | 0.208 | -0.0158 | No | ||
| 30 | MTMR4 | 4540524 | 5056 | 0.186 | -0.0259 | No | ||
| 31 | UBE2C | 6130017 | 5204 | 0.173 | -0.0326 | No | ||
| 32 | MCM3 | 5570068 | 5213 | 0.172 | -0.0316 | No | ||
| 33 | PLK1 | 1780369 2640121 | 5389 | 0.159 | -0.0399 | No | ||
| 34 | B4GALT5 | 1230692 | 5672 | 0.138 | -0.0541 | No | ||
| 35 | CTGF | 4540577 | 5729 | 0.135 | -0.0561 | No | ||
| 36 | NUFIP1 | 6350241 | 6343 | 0.102 | -0.0884 | No | ||
| 37 | HS3ST3B1 | 380040 | 6470 | 0.096 | -0.0945 | No | ||
| 38 | RNF14 | 50402 1340041 6370687 | 6718 | 0.087 | -0.1072 | No | ||
| 39 | HSPA1L | 4010538 | 7122 | 0.073 | -0.1285 | No | ||
| 40 | TIAM2 | 360195 | 7124 | 0.072 | -0.1279 | No | ||
| 41 | BIRC6 | 70148 | 7578 | 0.060 | -0.1520 | No | ||
| 42 | CDH11 | 1230133 2100180 4610110 | 7613 | 0.059 | -0.1534 | No | ||
| 43 | CDK7 | 2640451 | 7739 | 0.056 | -0.1597 | No | ||
| 44 | UBN1 | 5900195 | 7928 | 0.051 | -0.1695 | No | ||
| 45 | IPO7 | 2190746 | 8346 | 0.041 | -0.1917 | No | ||
| 46 | CXXC5 | 840010 | 8406 | 0.040 | -0.1946 | No | ||
| 47 | RAB5A | 6520324 | 8472 | 0.039 | -0.1978 | No | ||
| 48 | CDC42BPA | 840671 | 8865 | 0.031 | -0.2188 | No | ||
| 49 | CRSP6 | 2940358 | 8953 | 0.029 | -0.2233 | No | ||
| 50 | FOSL1 | 430021 | 8986 | 0.029 | -0.2248 | No | ||
| 51 | EPS8 | 7050204 | 9057 | 0.028 | -0.2284 | No | ||
| 52 | KIF11 | 5390139 | 9245 | 0.025 | -0.2383 | No | ||
| 53 | FZD1 | 3140215 | 9539 | 0.020 | -0.2540 | No | ||
| 54 | MFAP1 | 6660092 | 9810 | 0.015 | -0.2685 | No | ||
| 55 | BUB1 | 5390270 | 10159 | 0.009 | -0.2873 | No | ||
| 56 | CFLAR | 2320605 6180440 | 10607 | 0.002 | -0.3115 | No | ||
| 57 | ATG5 | 6200433 5360324 | 11032 | -0.005 | -0.3344 | No | ||
| 58 | FBXL7 | 6620711 | 11362 | -0.010 | -0.3521 | No | ||
| 59 | NMI | 520270 | 11445 | -0.012 | -0.3565 | No | ||
| 60 | NCOA6 | 1780333 6450110 | 11461 | -0.012 | -0.3572 | No | ||
| 61 | VEGFC | 5910494 | 11571 | -0.014 | -0.3630 | No | ||
| 62 | MAPKAPK5 | 2510601 | 11592 | -0.015 | -0.3639 | No | ||
| 63 | TDG | 2900358 2940079 | 11708 | -0.017 | -0.3700 | No | ||
| 64 | PLAG1 | 1450142 3870139 6520039 | 11711 | -0.017 | -0.3700 | No | ||
| 65 | MTIF2 | 2260270 | 12648 | -0.038 | -0.4204 | No | ||
| 66 | TCERG1 | 2350551 4540397 5220746 | 13273 | -0.059 | -0.4537 | No | ||
| 67 | NBN | 730538 2470619 4780594 | 13322 | -0.060 | -0.4558 | No | ||
| 68 | TRIP4 | 630072 | 13408 | -0.064 | -0.4599 | No | ||
| 69 | CDC23 | 3190593 | 13459 | -0.066 | -0.4621 | No | ||
| 70 | RBM27 | 4050603 | 13474 | -0.067 | -0.4624 | No | ||
| 71 | SFRS9 | 3800047 | 14243 | -0.112 | -0.5031 | No | ||
| 72 | JMJD1C | 940575 2120025 | 14253 | -0.113 | -0.5027 | No | ||
| 73 | CSNK2A1 | 1580577 | 14287 | -0.116 | -0.5035 | No | ||
| 74 | ATF1 | 4570142 5550095 | 14358 | -0.123 | -0.5064 | No | ||
| 75 | CYHR1 | 940609 2810463 3190332 3290692 4280411 | 14447 | -0.130 | -0.5101 | No | ||
| 76 | PSMC2 | 7040010 | 14507 | -0.135 | -0.5123 | No | ||
| 77 | SSFA2 | 3870133 | 14546 | -0.139 | -0.5133 | No | ||
| 78 | GOPC | 6620400 | 14775 | -0.168 | -0.5243 | No | ||
| 79 | CDC5L | 3130446 3390025 6400204 | 14845 | -0.177 | -0.5266 | No | ||
| 80 | USP12 | 6130735 6620280 | 15102 | -0.221 | -0.5388 | No | ||
| 81 | PAFAH2 | 3140451 5720593 6510014 | 15176 | -0.235 | -0.5409 | No | ||
| 82 | FNDC3A | 2690097 3290044 | 15178 | -0.236 | -0.5391 | No | ||
| 83 | PPP1CC | 6380300 2510647 | 15240 | -0.251 | -0.5405 | No | ||
| 84 | PSMB1 | 2940402 | 15530 | -0.317 | -0.5537 | No | ||
| 85 | MRPL15 | 360390 | 15720 | -0.375 | -0.5610 | No | ||
| 86 | FBXL3 | 5910142 | 15764 | -0.387 | -0.5603 | No | ||
| 87 | SMARCA5 | 6620050 | 15842 | -0.416 | -0.5612 | No | ||
| 88 | GTF2H1 | 2570746 2650148 | 15865 | -0.423 | -0.5591 | No | ||
| 89 | PRIM2A | 7000692 | 15959 | -0.456 | -0.5606 | No | ||
| 90 | BCL2L1 | 1580452 4200152 5420484 | 16084 | -0.495 | -0.5635 | Yes | ||
| 91 | PSME3 | 2810537 | 16141 | -0.515 | -0.5625 | Yes | ||
| 92 | GABPB2 | 6130450 7050735 | 16200 | -0.538 | -0.5615 | Yes | ||
| 93 | ERCC3 | 6900008 | 16234 | -0.551 | -0.5590 | Yes | ||
| 94 | PSMC6 | 2810021 | 16260 | -0.560 | -0.5560 | Yes | ||
| 95 | RCOR1 | 5360717 | 16331 | -0.587 | -0.5552 | Yes | ||
| 96 | ZDHHC5 | 1690711 | 16359 | -0.598 | -0.5520 | Yes | ||
| 97 | CCNA2 | 5290075 | 16440 | -0.634 | -0.5514 | Yes | ||
| 98 | CRLF3 | 6620019 | 16549 | -0.685 | -0.5520 | Yes | ||
| 99 | BIRC2 | 2940435 4730020 | 16663 | -0.757 | -0.5522 | Yes | ||
| 100 | STK17B | 130091 | 16859 | -0.874 | -0.5560 | Yes | ||
| 101 | EML4 | 5700373 | 16965 | -0.941 | -0.5543 | Yes | ||
| 102 | MAT2A | 4070026 4730079 6020280 | 17061 | -0.995 | -0.5517 | Yes | ||
| 103 | TXNRD1 | 6590446 | 17245 | -1.098 | -0.5531 | Yes | ||
| 104 | UBE2D2 | 4590671 6510446 | 17253 | -1.102 | -0.5449 | Yes | ||
| 105 | MAPK14 | 5290731 | 17315 | -1.150 | -0.5393 | Yes | ||
| 106 | SUPV3L1 | 1170072 | 17329 | -1.165 | -0.5309 | Yes | ||
| 107 | SIRT1 | 1190731 | 17371 | -1.201 | -0.5238 | Yes | ||
| 108 | HSPH1 | 1690017 | 17447 | -1.265 | -0.5180 | Yes | ||
| 109 | RBM17 | 870592 | 17448 | -1.265 | -0.5082 | Yes | ||
| 110 | GTF2B | 6860685 | 17511 | -1.328 | -0.5012 | Yes | ||
| 111 | TMPO | 4050494 | 17673 | -1.489 | -0.4984 | Yes | ||
| 112 | XBP1 | 3840594 | 17677 | -1.491 | -0.4870 | Yes | ||
| 113 | TOP2A | 360717 | 17707 | -1.531 | -0.4766 | Yes | ||
| 114 | ELOVL6 | 5340746 | 17800 | -1.633 | -0.4689 | Yes | ||
| 115 | OSBPL9 | 2570373 | 17888 | -1.715 | -0.4603 | Yes | ||
| 116 | MCTS1 | 6130133 | 17980 | -1.821 | -0.4511 | Yes | ||
| 117 | TNFSF5IP1 | 2060095 | 18012 | -1.858 | -0.4383 | Yes | ||
| 118 | RAP1B | 3710138 | 18017 | -1.861 | -0.4240 | Yes | ||
| 119 | POLD3 | 6400278 | 18063 | -1.899 | -0.4117 | Yes | ||
| 120 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.3974 | Yes | ||
| 121 | MAP3K7 | 6040068 | 18082 | -1.929 | -0.3828 | Yes | ||
| 122 | PCNA | 940754 | 18090 | -1.941 | -0.3681 | Yes | ||
| 123 | PJA2 | 430537 | 18146 | -2.037 | -0.3552 | Yes | ||
| 124 | GTF2H2 | 5130358 | 18171 | -2.069 | -0.3404 | Yes | ||
| 125 | DDX21 | 6100446 | 18174 | -2.073 | -0.3244 | Yes | ||
| 126 | FNTA | 70059 | 18207 | -2.120 | -0.3097 | Yes | ||
| 127 | HDAC2 | 4050433 | 18244 | -2.188 | -0.2946 | Yes | ||
| 128 | SLC25A17 | 2100369 | 18246 | -2.189 | -0.2777 | Yes | ||
| 129 | TBK1 | 5690338 | 18268 | -2.222 | -0.2615 | Yes | ||
| 130 | PRKACB | 4210170 | 18277 | -2.235 | -0.2446 | Yes | ||
| 131 | RIPK1 | 3190025 | 18291 | -2.264 | -0.2277 | Yes | ||
| 132 | CCNH | 3190347 | 18307 | -2.289 | -0.2107 | Yes | ||
| 133 | RPP14 | 7040014 | 18382 | -2.470 | -0.1955 | Yes | ||
| 134 | PSMA4 | 4560427 | 18418 | -2.539 | -0.1777 | Yes | ||
| 135 | DDX20 | 3290348 | 18454 | -2.649 | -0.1590 | Yes | ||
| 136 | RABGGTB | 3120037 | 18464 | -2.678 | -0.1387 | Yes | ||
| 137 | RAB1A | 2370671 | 18476 | -2.727 | -0.1181 | Yes | ||
| 138 | PSMD14 | 5690593 | 18497 | -2.806 | -0.0974 | Yes | ||
| 139 | OGT | 2360131 4610333 | 18507 | -2.829 | -0.0759 | Yes | ||
| 140 | CCT4 | 6380161 | 18545 | -3.052 | -0.0541 | Yes | ||
| 141 | LBR | 4540671 | 18590 | -3.582 | -0.0287 | Yes | ||
| 142 | SLC4A1AP | 3190576 | 18602 | -3.866 | 0.0008 | Yes |

