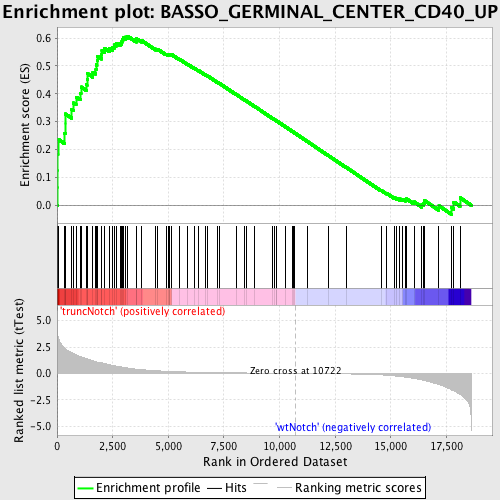
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | truncNotch |
| GeneSet | BASSO_GERMINAL_CENTER_CD40_UP |
| Enrichment Score (ES) | 0.60716563 |
| Normalized Enrichment Score (NES) | 1.6333139 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15555364 |
| FWER p-Value | 0.873 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BATF | 6760390 | 19 | 4.143 | 0.0629 | Yes | ||
| 2 | NFKBIE | 580390 2190086 | 22 | 4.000 | 0.1245 | Yes | ||
| 3 | MAPKAPK2 | 2850500 | 26 | 3.846 | 0.1837 | Yes | ||
| 4 | PTK2B | 4730411 | 40 | 3.507 | 0.2371 | Yes | ||
| 5 | PIK3CD | 3060546 | 340 | 2.382 | 0.2577 | Yes | ||
| 6 | LHFPL2 | 450020 | 371 | 2.327 | 0.2920 | Yes | ||
| 7 | TAP1 | 4050047 | 372 | 2.324 | 0.3279 | Yes | ||
| 8 | UNC119 | 360372 | 657 | 1.963 | 0.3428 | Yes | ||
| 9 | RAC2 | 1580541 | 738 | 1.867 | 0.3673 | Yes | ||
| 10 | IFI30 | 730309 2940500 | 855 | 1.748 | 0.3880 | Yes | ||
| 11 | SYNGR2 | 4050332 4560091 6130348 | 1043 | 1.585 | 0.4024 | Yes | ||
| 12 | CCR7 | 2060687 | 1076 | 1.557 | 0.4247 | Yes | ||
| 13 | EBI2 | 6220722 | 1326 | 1.383 | 0.4326 | Yes | ||
| 14 | PTP4A3 | 3780504 6400364 | 1347 | 1.371 | 0.4527 | Yes | ||
| 15 | CCL4 | 50368 430047 | 1374 | 1.353 | 0.4721 | Yes | ||
| 16 | IRF5 | 3360519 | 1606 | 1.203 | 0.4782 | Yes | ||
| 17 | EHD1 | 3840731 | 1743 | 1.117 | 0.4881 | Yes | ||
| 18 | NFKBIA | 1570152 | 1757 | 1.101 | 0.5044 | Yes | ||
| 19 | CAPG | 770373 | 1793 | 1.079 | 0.5192 | Yes | ||
| 20 | NFKB2 | 2320670 | 1817 | 1.069 | 0.5344 | Yes | ||
| 21 | STK10 | 2760010 3940605 | 1997 | 0.995 | 0.5401 | Yes | ||
| 22 | IER3 | 5860021 | 2014 | 0.990 | 0.5545 | Yes | ||
| 23 | LITAF | 6940671 | 2132 | 0.932 | 0.5626 | Yes | ||
| 24 | STAT5A | 2680458 | 2347 | 0.833 | 0.5639 | Yes | ||
| 25 | GADD45B | 2350408 | 2500 | 0.766 | 0.5675 | Yes | ||
| 26 | CD83 | 4150242 | 2563 | 0.739 | 0.5756 | Yes | ||
| 27 | ISGF3G | 3190131 7100142 | 2649 | 0.696 | 0.5817 | Yes | ||
| 28 | LAPTM5 | 5550372 | 2834 | 0.622 | 0.5814 | Yes | ||
| 29 | TRIP10 | 460438 | 2910 | 0.596 | 0.5866 | Yes | ||
| 30 | TMC6 | 60736 1230093 2100288 4850735 | 2922 | 0.592 | 0.5951 | Yes | ||
| 31 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 2962 | 0.579 | 0.6019 | Yes | ||
| 32 | ADAM8 | 450347 5670609 | 3077 | 0.535 | 0.6040 | Yes | ||
| 33 | JUNB | 4230048 | 3164 | 0.504 | 0.6072 | Yes | ||
| 34 | TNFAIP3 | 2900142 | 3560 | 0.405 | 0.5921 | No | ||
| 35 | DDIT4 | 2570408 | 3564 | 0.404 | 0.5982 | No | ||
| 36 | TNFAIP2 | 70438 1740215 | 3795 | 0.354 | 0.5912 | No | ||
| 37 | HTR3A | 3310180 | 4429 | 0.254 | 0.5610 | No | ||
| 38 | SH2B3 | 4810451 4850128 | 4528 | 0.240 | 0.5594 | No | ||
| 39 | CR2 | 2850332 6450397 | 4897 | 0.202 | 0.5427 | No | ||
| 40 | CCL3 | 2810092 | 5023 | 0.190 | 0.5389 | No | ||
| 41 | RGS1 | 4060347 4540181 | 5036 | 0.188 | 0.5411 | No | ||
| 42 | MTMR4 | 4540524 | 5056 | 0.186 | 0.5430 | No | ||
| 43 | ICOSLG | 70128 | 5145 | 0.178 | 0.5410 | No | ||
| 44 | ARID5B | 110403 | 5496 | 0.151 | 0.5244 | No | ||
| 45 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 5857 | 0.127 | 0.5069 | No | ||
| 46 | TCFL5 | 2190019 | 6180 | 0.110 | 0.4913 | No | ||
| 47 | CD40 | 2760471 2940390 6220402 6220452 | 6358 | 0.101 | 0.4833 | No | ||
| 48 | TNFSF9 | 1190408 | 6666 | 0.089 | 0.4681 | No | ||
| 49 | LTA | 1740088 | 6769 | 0.085 | 0.4639 | No | ||
| 50 | MAP3K8 | 2940286 | 7203 | 0.070 | 0.4416 | No | ||
| 51 | MEF2C | 670025 780338 | 7314 | 0.067 | 0.4367 | No | ||
| 52 | SNF1LK | 6110403 | 8053 | 0.048 | 0.3976 | No | ||
| 53 | PLXNC1 | 5860315 | 8437 | 0.040 | 0.3776 | No | ||
| 54 | ARHGEF2 | 3360577 | 8524 | 0.038 | 0.3735 | No | ||
| 55 | PLEK | 1090039 | 8866 | 0.031 | 0.3556 | No | ||
| 56 | NCF2 | 540129 2370441 2650133 | 9688 | 0.017 | 0.3116 | No | ||
| 57 | MYB | 1660494 5860451 6130706 | 9771 | 0.015 | 0.3074 | No | ||
| 58 | TMSB4X | 6620114 | 9838 | 0.014 | 0.3040 | No | ||
| 59 | TRAF1 | 3440735 | 9843 | 0.014 | 0.3040 | No | ||
| 60 | IRF4 | 610133 | 10268 | 0.007 | 0.2813 | No | ||
| 61 | ELL2 | 6420091 | 10575 | 0.002 | 0.2648 | No | ||
| 62 | CFLAR | 2320605 6180440 | 10607 | 0.002 | 0.2632 | No | ||
| 63 | ZFP36L1 | 2510138 4120048 | 10657 | 0.001 | 0.2605 | No | ||
| 64 | CCL22 | 6380086 | 11269 | -0.009 | 0.2277 | No | ||
| 65 | PHACTR1 | 3190176 5670286 | 12198 | -0.027 | 0.1781 | No | ||
| 66 | LYN | 6040600 | 13008 | -0.048 | 0.1352 | No | ||
| 67 | RXRA | 3800471 | 14563 | -0.141 | 0.0535 | No | ||
| 68 | SLAMF1 | 580131 4920110 | 14809 | -0.173 | 0.0429 | No | ||
| 69 | DUSP2 | 2370184 | 15174 | -0.235 | 0.0269 | No | ||
| 70 | BLR1 | 1500300 5420017 | 15275 | -0.256 | 0.0255 | No | ||
| 71 | IL1B | 2640364 | 15409 | -0.286 | 0.0227 | No | ||
| 72 | DTX2 | 4210041 7000008 | 15512 | -0.312 | 0.0220 | No | ||
| 73 | KCNN4 | 2680129 | 15643 | -0.349 | 0.0204 | No | ||
| 74 | TCEB3 | 5890156 | 15702 | -0.368 | 0.0230 | No | ||
| 75 | STAT1 | 6510204 6590553 | 16041 | -0.480 | 0.0121 | No | ||
| 76 | LGALS1 | 6450193 | 16396 | -0.613 | 0.0025 | No | ||
| 77 | TNF | 6650603 | 16480 | -0.652 | 0.0081 | No | ||
| 78 | RAB27A | 450019 | 16522 | -0.673 | 0.0162 | No | ||
| 79 | ICAM1 | 6980138 | 17164 | -1.039 | -0.0023 | No | ||
| 80 | CHD1 | 4590048 | 17736 | -1.565 | -0.0090 | No | ||
| 81 | HSP90B1 | 5670576 | 17831 | -1.665 | 0.0117 | No | ||
| 82 | TNFAIP8 | 2760551 | 18112 | -1.986 | 0.0272 | No |

