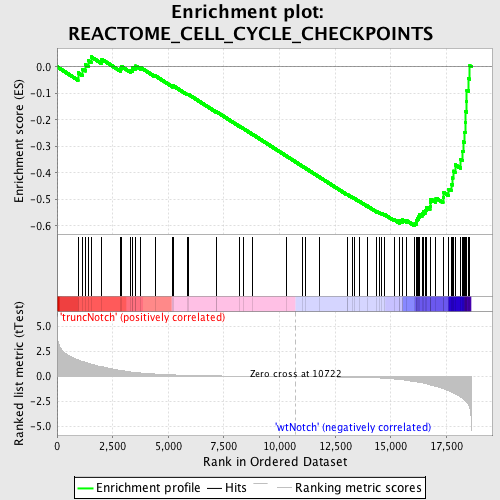
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | -0.6008194 |
| Normalized Enrichment Score (NES) | -1.7061964 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15363324 |
| FWER p-Value | 0.472 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMD2 | 4670706 5050364 | 941 | 1.679 | -0.0227 | No | ||
| 2 | PSMA1 | 380059 2760195 | 1149 | 1.500 | -0.0088 | No | ||
| 3 | PSMC1 | 6350538 | 1283 | 1.414 | 0.0076 | No | ||
| 4 | PSMD9 | 50020 5890368 | 1407 | 1.336 | 0.0233 | No | ||
| 5 | RFC5 | 3800452 6020091 | 1531 | 1.251 | 0.0375 | No | ||
| 6 | PSMA7 | 2230131 | 2006 | 0.993 | 0.0285 | No | ||
| 7 | PSMB6 | 2690711 | 2861 | 0.612 | -0.0073 | No | ||
| 8 | PSMB10 | 630504 6510039 | 2884 | 0.604 | 0.0016 | No | ||
| 9 | MCM2 | 5050139 | 3319 | 0.459 | -0.0142 | No | ||
| 10 | TP53 | 6130707 | 3380 | 0.442 | -0.0100 | No | ||
| 11 | BUB3 | 3170546 | 3396 | 0.440 | -0.0035 | No | ||
| 12 | CDC20 | 3440017 3440044 6220088 | 3513 | 0.413 | -0.0028 | No | ||
| 13 | MCM5 | 2680647 | 3531 | 0.411 | 0.0031 | No | ||
| 14 | PSMB3 | 4280594 | 3764 | 0.359 | -0.0034 | No | ||
| 15 | PSMA5 | 5390537 | 4404 | 0.258 | -0.0335 | No | ||
| 16 | UBE2C | 6130017 | 5204 | 0.173 | -0.0738 | No | ||
| 17 | MCM3 | 5570068 | 5213 | 0.172 | -0.0713 | No | ||
| 18 | PSMB4 | 520402 | 5847 | 0.127 | -0.1033 | No | ||
| 19 | RAD1 | 4200551 | 5893 | 0.125 | -0.1037 | No | ||
| 20 | RFC3 | 1980600 | 7145 | 0.072 | -0.1699 | No | ||
| 21 | PSMB9 | 6980471 | 7156 | 0.072 | -0.1693 | No | ||
| 22 | PSMA3 | 5900047 7040161 | 8176 | 0.045 | -0.2235 | No | ||
| 23 | PSMC3 | 2480184 | 8367 | 0.041 | -0.2330 | No | ||
| 24 | PSMD5 | 3940139 4570041 | 8769 | 0.033 | -0.2541 | No | ||
| 25 | PSMA6 | 50609 | 10307 | 0.006 | -0.3369 | No | ||
| 26 | PSMD8 | 630142 | 11013 | -0.005 | -0.3748 | No | ||
| 27 | UBE2D1 | 1850400 | 11164 | -0.007 | -0.3828 | No | ||
| 28 | CDKN1A | 4050088 6400706 | 11801 | -0.019 | -0.4168 | No | ||
| 29 | CCNB2 | 6510528 | 13046 | -0.050 | -0.4831 | No | ||
| 30 | PSMB5 | 6290242 | 13050 | -0.050 | -0.4824 | No | ||
| 31 | CHEK1 | 50167 1940300 | 13289 | -0.059 | -0.4942 | No | ||
| 32 | MCM10 | 4920632 | 13362 | -0.062 | -0.4971 | No | ||
| 33 | PSMD11 | 2340538 6510053 | 13588 | -0.072 | -0.5080 | No | ||
| 34 | ATM | 3610110 4050524 | 13930 | -0.090 | -0.5249 | No | ||
| 35 | CDC25C | 2570673 4760161 6520707 | 14375 | -0.124 | -0.5468 | No | ||
| 36 | PSMC2 | 7040010 | 14507 | -0.135 | -0.5516 | No | ||
| 37 | ORC2L | 1990470 6510019 | 14587 | -0.144 | -0.5534 | No | ||
| 38 | WEE1 | 3390070 | 14732 | -0.160 | -0.5585 | No | ||
| 39 | ORC1L | 2370328 6110390 | 15143 | -0.230 | -0.5768 | No | ||
| 40 | RPA2 | 2760301 5420195 | 15395 | -0.283 | -0.5856 | No | ||
| 41 | CDK2 | 130484 2260301 4010088 5050110 | 15400 | -0.284 | -0.5811 | No | ||
| 42 | PSMB2 | 940035 4210324 | 15519 | -0.313 | -0.5822 | No | ||
| 43 | PSMB1 | 2940402 | 15530 | -0.317 | -0.5775 | No | ||
| 44 | MDM2 | 3450053 5080138 | 15723 | -0.375 | -0.5816 | No | ||
| 45 | ORC4L | 4230538 5550288 | 16081 | -0.494 | -0.5926 | Yes | ||
| 46 | PSME3 | 2810537 | 16141 | -0.515 | -0.5872 | Yes | ||
| 47 | PSMC4 | 580050 1580025 | 16153 | -0.518 | -0.5791 | Yes | ||
| 48 | HUS1 | 3440494 3800156 | 16187 | -0.533 | -0.5720 | Yes | ||
| 49 | PSMC6 | 2810021 | 16260 | -0.560 | -0.5665 | Yes | ||
| 50 | MCM7 | 3290292 5220056 | 16282 | -0.569 | -0.5582 | Yes | ||
| 51 | CDC25A | 3800184 | 16404 | -0.617 | -0.5544 | Yes | ||
| 52 | PSMC5 | 2760315 6550021 | 16473 | -0.650 | -0.5472 | Yes | ||
| 53 | CDC45L | 70537 3130114 | 16566 | -0.690 | -0.5407 | Yes | ||
| 54 | RAD9B | 2320064 | 16609 | -0.722 | -0.5309 | Yes | ||
| 55 | PSMD3 | 1400647 | 16762 | -0.811 | -0.5255 | Yes | ||
| 56 | PSME1 | 450193 4480035 | 16780 | -0.823 | -0.5127 | Yes | ||
| 57 | MCM6 | 60092 540181 6510110 | 16794 | -0.831 | -0.4995 | Yes | ||
| 58 | CDC16 | 1940706 | 17027 | -0.980 | -0.4957 | Yes | ||
| 59 | UBE2E1 | 4230064 1090440 | 17347 | -1.186 | -0.4931 | Yes | ||
| 60 | PSMD4 | 430068 | 17380 | -1.214 | -0.4745 | Yes | ||
| 61 | ORC5L | 1940133 1940711 | 17603 | -1.414 | -0.4629 | Yes | ||
| 62 | MCM4 | 2760673 5420711 | 17712 | -1.538 | -0.4430 | Yes | ||
| 63 | PSMD10 | 520494 1170576 3830050 | 17774 | -1.611 | -0.4194 | Yes | ||
| 64 | RPA3 | 5700136 | 17795 | -1.631 | -0.3933 | Yes | ||
| 65 | CDC7 | 4060546 4850041 | 17891 | -1.718 | -0.3697 | Yes | ||
| 66 | PSMD12 | 730044 | 18128 | -2.007 | -0.3489 | Yes | ||
| 67 | RPA1 | 360452 | 18231 | -2.159 | -0.3184 | Yes | ||
| 68 | PSMA2 | 6510093 | 18263 | -2.215 | -0.2831 | Yes | ||
| 69 | PSMD7 | 2030619 6220594 | 18323 | -2.337 | -0.2472 | Yes | ||
| 70 | MAD2L1 | 4480725 | 18343 | -2.379 | -0.2086 | Yes | ||
| 71 | BUB1B | 1450288 | 18355 | -2.413 | -0.1689 | Yes | ||
| 72 | CCNB1 | 4590433 4780372 | 18405 | -2.518 | -0.1295 | Yes | ||
| 73 | PSMA4 | 4560427 | 18418 | -2.539 | -0.0877 | Yes | ||
| 74 | PSMD14 | 5690593 | 18497 | -2.806 | -0.0451 | Yes | ||
| 75 | RFC4 | 3800082 6840142 | 18552 | -3.081 | 0.0035 | Yes |

