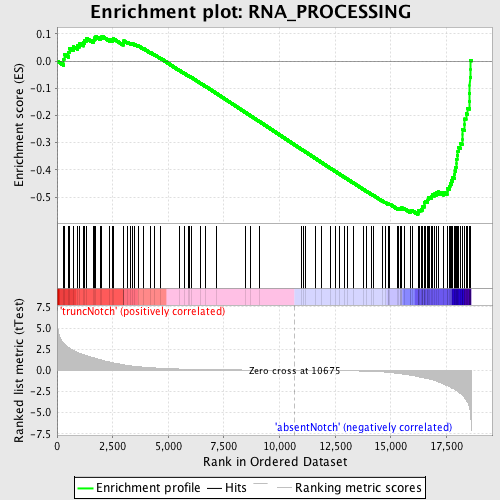
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | -0.5641964 |
| Normalized Enrichment Score (NES) | -1.6240301 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.3623528 |
| FWER p-Value | 0.908 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SF3A2 | 3140500 | 301 | 3.190 | 0.0047 | No | ||
| 2 | GEMIN7 | 2320433 | 312 | 3.146 | 0.0249 | No | ||
| 3 | DDX54 | 6220324 | 512 | 2.688 | 0.0318 | No | ||
| 4 | IGF2BP3 | 2810161 4230338 | 544 | 2.645 | 0.0476 | No | ||
| 5 | LSM4 | 3130180 | 741 | 2.342 | 0.0524 | No | ||
| 6 | SNRPB | 1230097 2940086 3520671 | 921 | 2.104 | 0.0566 | No | ||
| 7 | METTL1 | 4540670 5860162 7040433 | 1018 | 2.003 | 0.0646 | No | ||
| 8 | RBPMS | 3990494 | 1193 | 1.837 | 0.0673 | No | ||
| 9 | GEMIN5 | 3120577 | 1231 | 1.794 | 0.0771 | No | ||
| 10 | UPF1 | 430164 1190750 | 1306 | 1.735 | 0.0845 | No | ||
| 11 | SNRPA1 | 1050735 6110131 | 1624 | 1.494 | 0.0772 | No | ||
| 12 | SF3A1 | 1580315 | 1663 | 1.472 | 0.0848 | No | ||
| 13 | SNRP70 | 4200181 | 1742 | 1.416 | 0.0899 | No | ||
| 14 | SFRS2 | 50707 380593 | 1955 | 1.237 | 0.0866 | No | ||
| 15 | U2AF1 | 5130292 | 2011 | 1.199 | 0.0915 | No | ||
| 16 | SFRS10 | 6130528 6400438 7040112 | 2363 | 0.992 | 0.0791 | No | ||
| 17 | SNRPD2 | 6110035 | 2476 | 0.923 | 0.0791 | No | ||
| 18 | SFRS9 | 3800047 | 2527 | 0.890 | 0.0823 | No | ||
| 19 | EXOSC2 | 6900471 | 2964 | 0.666 | 0.0631 | No | ||
| 20 | RBMY1A1 | 6200053 | 2994 | 0.650 | 0.0658 | No | ||
| 21 | SFRS8 | 5130162 5700164 | 2996 | 0.650 | 0.0700 | No | ||
| 22 | RRP9 | 630451 | 3003 | 0.648 | 0.0740 | No | ||
| 23 | DHX16 | 940044 | 3178 | 0.582 | 0.0684 | No | ||
| 24 | DDX39 | 3840148 | 3294 | 0.541 | 0.0657 | No | ||
| 25 | RBM6 | 840279 3840563 4780129 6020446 | 3373 | 0.514 | 0.0649 | No | ||
| 26 | SRRM1 | 6110025 | 3498 | 0.469 | 0.0613 | No | ||
| 27 | EXOSC3 | 1740064 | 3638 | 0.432 | 0.0566 | No | ||
| 28 | NONO | 7050014 | 3881 | 0.377 | 0.0460 | No | ||
| 29 | PTBP1 | 4070736 | 4215 | 0.315 | 0.0301 | No | ||
| 30 | SF3B4 | 110072 | 4390 | 0.286 | 0.0226 | No | ||
| 31 | BCAS2 | 2340494 | 4652 | 0.247 | 0.0101 | No | ||
| 32 | ADAT1 | 510390 6900019 | 5483 | 0.161 | -0.0338 | No | ||
| 33 | HNRPUL1 | 2120008 4920408 5570180 6370609 6770403 | 5720 | 0.143 | -0.0456 | No | ||
| 34 | PPAN | 540398 | 5922 | 0.129 | -0.0556 | No | ||
| 35 | RNPS1 | 610736 1090021 | 5951 | 0.127 | -0.0563 | No | ||
| 36 | INTS6 | 6040736 7570767 | 6019 | 0.122 | -0.0591 | No | ||
| 37 | NUFIP1 | 6350241 | 6438 | 0.100 | -0.0810 | No | ||
| 38 | FARS2 | 5340575 | 6664 | 0.091 | -0.0926 | No | ||
| 39 | CUGBP1 | 450292 510022 7050176 7050215 | 7149 | 0.073 | -0.1183 | No | ||
| 40 | LSM5 | 4540398 | 8480 | 0.040 | -0.1899 | No | ||
| 41 | SSB | 460286 3140717 | 8687 | 0.035 | -0.2008 | No | ||
| 42 | NOL3 | 4060605 4670121 6620487 | 9088 | 0.027 | -0.2223 | No | ||
| 43 | SF3B3 | 3710044 | 10980 | -0.005 | -0.3245 | No | ||
| 44 | CPSF3 | 2480671 | 11058 | -0.006 | -0.3286 | No | ||
| 45 | GEMIN6 | 2120288 3830176 | 11185 | -0.009 | -0.3354 | No | ||
| 46 | SLBP | 6220601 | 11603 | -0.016 | -0.3578 | No | ||
| 47 | HNRPF | 670138 | 11877 | -0.022 | -0.3724 | No | ||
| 48 | SNRPB2 | 5720577 | 12278 | -0.031 | -0.3939 | No | ||
| 49 | SYNCRIP | 1690195 3140113 4670279 | 12501 | -0.036 | -0.4056 | No | ||
| 50 | PRPF4B | 940181 2570300 | 12711 | -0.042 | -0.4166 | No | ||
| 51 | SNRPN | 4670195 5890494 6620110 | 12898 | -0.048 | -0.4264 | No | ||
| 52 | BICD1 | 110128 2940324 | 13048 | -0.053 | -0.4341 | No | ||
| 53 | PABPC4 | 1990170 6760270 5390138 | 13329 | -0.063 | -0.4488 | No | ||
| 54 | CSTF3 | 3850156 5570458 | 13768 | -0.086 | -0.4719 | No | ||
| 55 | DHX8 | 6100139 | 13917 | -0.096 | -0.4793 | No | ||
| 56 | HNRPD | 4120021 | 14130 | -0.114 | -0.4900 | No | ||
| 57 | PPARGC1A | 4670040 | 14233 | -0.125 | -0.4947 | No | ||
| 58 | RPS14 | 430541 | 14613 | -0.177 | -0.5140 | No | ||
| 59 | TXNL4A | 4120286 6510242 | 14746 | -0.204 | -0.5198 | No | ||
| 60 | NUDT21 | 1240167 | 14876 | -0.230 | -0.5253 | No | ||
| 61 | ZNF346 | 2650577 3990170 4150348 | 14890 | -0.232 | -0.5245 | No | ||
| 62 | CRNKL1 | 5050097 | 14951 | -0.248 | -0.5261 | No | ||
| 63 | ELAVL4 | 50735 3360086 5220167 | 15306 | -0.352 | -0.5429 | No | ||
| 64 | NOLA2 | 4060167 | 15338 | -0.365 | -0.5422 | No | ||
| 65 | GEMIN4 | 130278 2230075 | 15427 | -0.396 | -0.5443 | No | ||
| 66 | PRPF31 | 6100360 | 15442 | -0.399 | -0.5424 | No | ||
| 67 | FUSIP1 | 520082 5390114 | 15473 | -0.410 | -0.5414 | No | ||
| 68 | PRPF3 | 2510435 2940593 3610113 5390722 | 15493 | -0.417 | -0.5397 | No | ||
| 69 | SNRPF | 3170280 | 15621 | -0.466 | -0.5434 | No | ||
| 70 | FBL | 5130020 | 15878 | -0.566 | -0.5536 | No | ||
| 71 | PRPF8 | 1230497 1570600 | 15902 | -0.578 | -0.5510 | No | ||
| 72 | SNW1 | 4010736 | 15957 | -0.608 | -0.5499 | No | ||
| 73 | NCBP2 | 5340500 | 16222 | -0.747 | -0.5593 | Yes | ||
| 74 | RNGTT | 780152 780746 1090471 | 16226 | -0.749 | -0.5545 | Yes | ||
| 75 | PHF5A | 2690519 | 16239 | -0.753 | -0.5502 | Yes | ||
| 76 | SFRS5 | 3450176 6350008 | 16289 | -0.778 | -0.5477 | Yes | ||
| 77 | CSTF1 | 4610129 | 16358 | -0.811 | -0.5461 | Yes | ||
| 78 | LSM3 | 780164 | 16408 | -0.841 | -0.5432 | Yes | ||
| 79 | SNRPD1 | 4480162 | 16420 | -0.847 | -0.5382 | Yes | ||
| 80 | EFTUD2 | 2470707 4610148 | 16438 | -0.861 | -0.5334 | Yes | ||
| 81 | SIP1 | 2640706 | 16501 | -0.896 | -0.5309 | Yes | ||
| 82 | DUSP11 | 1500088 4610280 | 16503 | -0.897 | -0.5250 | Yes | ||
| 83 | RNMT | 1770500 | 16504 | -0.898 | -0.5191 | Yes | ||
| 84 | EXOSC7 | 520538 | 16557 | -0.935 | -0.5158 | Yes | ||
| 85 | NOLC1 | 2350195 | 16652 | -0.983 | -0.5144 | Yes | ||
| 86 | AARS | 1570279 2370136 | 16670 | -0.992 | -0.5088 | Yes | ||
| 87 | RBM5 | 3520402 5270121 | 16681 | -0.998 | -0.5028 | Yes | ||
| 88 | SFRS7 | 2760408 | 16758 | -1.040 | -0.5000 | Yes | ||
| 89 | CPSF1 | 6290064 | 16841 | -1.090 | -0.4973 | Yes | ||
| 90 | USP39 | 1090242 3520435 4760736 | 16851 | -1.098 | -0.4905 | Yes | ||
| 91 | SNRPG | 2340551 | 16961 | -1.180 | -0.4887 | Yes | ||
| 92 | RBM3 | 2360739 5700167 | 17042 | -1.265 | -0.4847 | Yes | ||
| 93 | ZNF638 | 6510112 | 17142 | -1.369 | -0.4810 | Yes | ||
| 94 | ASCC3L1 | 6620494 | 17362 | -1.607 | -0.4822 | Yes | ||
| 95 | SF3B2 | 6590576 | 17529 | -1.789 | -0.4794 | Yes | ||
| 96 | SNRPD3 | 2970050 | 17532 | -1.795 | -0.4677 | Yes | ||
| 97 | PABPC1 | 2650180 2690253 6020632 1990270 | 17644 | -1.920 | -0.4611 | Yes | ||
| 98 | RBMS1 | 6400014 | 17662 | -1.950 | -0.4492 | Yes | ||
| 99 | DGCR8 | 3450632 | 17733 | -2.069 | -0.4393 | Yes | ||
| 100 | KIN | 6550014 | 17779 | -2.124 | -0.4278 | Yes | ||
| 101 | KHDRBS1 | 1240403 6040040 | 17875 | -2.247 | -0.4181 | Yes | ||
| 102 | SRPK1 | 450110 | 17883 | -2.264 | -0.4036 | Yes | ||
| 103 | IVNS1ABP | 4760601 6520113 | 17911 | -2.312 | -0.3898 | Yes | ||
| 104 | TRNT1 | 870053 | 17937 | -2.350 | -0.3757 | Yes | ||
| 105 | NSUN2 | 2120193 | 17940 | -2.354 | -0.3603 | Yes | ||
| 106 | DHX38 | 780243 | 17977 | -2.415 | -0.3464 | Yes | ||
| 107 | HNRPDL | 1050102 1090181 5360471 | 18013 | -2.480 | -0.3319 | Yes | ||
| 108 | NOL5A | 5570139 | 18045 | -2.530 | -0.3170 | Yes | ||
| 109 | CSTF2 | 6040463 | 18125 | -2.727 | -0.3033 | Yes | ||
| 110 | SF3A3 | 6660603 | 18208 | -2.919 | -0.2885 | Yes | ||
| 111 | SFRS6 | 60224 | 18213 | -2.933 | -0.2694 | Yes | ||
| 112 | CDC2L2 | 1400671 5290168 | 18214 | -2.935 | -0.2501 | Yes | ||
| 113 | GRSF1 | 2100184 | 18310 | -3.204 | -0.2341 | Yes | ||
| 114 | SFRS2IP | 730687 1500373 | 18315 | -3.224 | -0.2131 | Yes | ||
| 115 | SFPQ | 4760110 | 18381 | -3.477 | -0.1937 | Yes | ||
| 116 | POP4 | 4560600 | 18455 | -3.785 | -0.1728 | Yes | ||
| 117 | SRPK2 | 6380341 | 18529 | -4.282 | -0.1485 | Yes | ||
| 118 | DDX20 | 3290348 | 18537 | -4.358 | -0.1202 | Yes | ||
| 119 | SPOP | 450035 | 18556 | -4.589 | -0.0909 | Yes | ||
| 120 | DBR1 | 1770286 | 18558 | -4.628 | -0.0605 | Yes | ||
| 121 | DHX15 | 870632 | 18572 | -4.807 | -0.0296 | Yes | ||
| 122 | SFRS1 | 2360440 | 18577 | -4.846 | 0.0021 | Yes |

