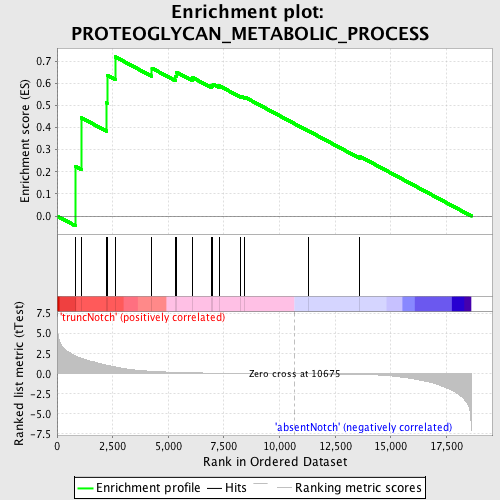
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | truncNotch |
| GeneSet | PROTEOGLYCAN_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.71877015 |
| Normalized Enrichment Score (NES) | 1.4347391 |
| Nominal p-value | 0.055172414 |
| FDR q-value | 0.984397 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | B4GALT7 | 870064 | 835 | 2.226 | 0.2242 | Yes | ||
| 2 | SGSH | 6180717 | 1087 | 1.929 | 0.4439 | Yes | ||
| 3 | CHST12 | 2030438 | 2224 | 1.062 | 0.5112 | Yes | ||
| 4 | HS6ST1 | 4760373 | 2250 | 1.046 | 0.6363 | Yes | ||
| 5 | GLCE | 4850040 | 2616 | 0.845 | 0.7188 | Yes | ||
| 6 | EXT1 | 4540603 | 4264 | 0.307 | 0.6673 | No | ||
| 7 | CHST11 | 6760546 | 5318 | 0.173 | 0.6316 | No | ||
| 8 | HS3ST3B1 | 380040 | 5372 | 0.169 | 0.6491 | No | ||
| 9 | NDST1 | 1500427 5340121 | 6091 | 0.119 | 0.6249 | No | ||
| 10 | CHST3 | 2370131 | 6931 | 0.080 | 0.5895 | No | ||
| 11 | CHST9 | 4560092 | 7006 | 0.077 | 0.5949 | No | ||
| 12 | LECT1 | 2640528 | 7289 | 0.069 | 0.5880 | No | ||
| 13 | IL17F | 670577 | 8252 | 0.044 | 0.5417 | No | ||
| 14 | CHST8 | 1940050 | 8428 | 0.041 | 0.5372 | No | ||
| 15 | CHST7 | 2760066 | 11302 | -0.011 | 0.3841 | No | ||
| 16 | HPSE | 6660021 | 13590 | -0.075 | 0.2702 | No |

