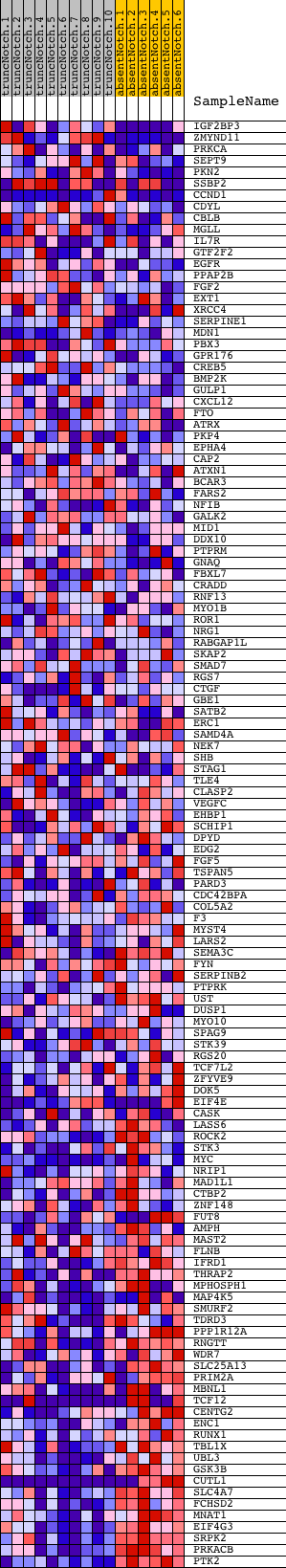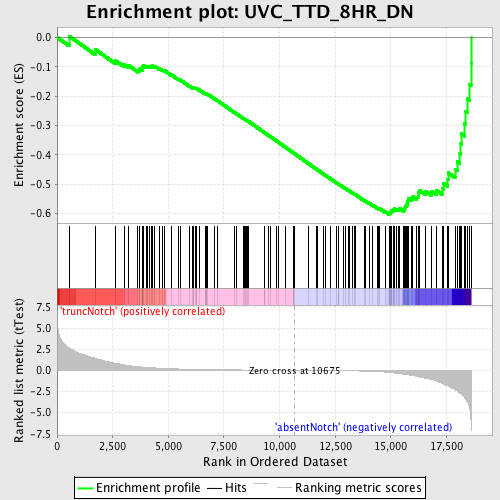
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | UVC_TTD_8HR_DN |
| Enrichment Score (ES) | -0.6027125 |
| Normalized Enrichment Score (NES) | -1.7448328 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.035880927 |
| FWER p-Value | 0.335 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGF2BP3 | 2810161 4230338 | 544 | 2.645 | 0.0042 | No | ||
| 2 | ZMYND11 | 630181 2570019 4850138 5360040 | 1725 | 1.433 | -0.0414 | No | ||
| 3 | PRKCA | 6400551 | 2627 | 0.839 | -0.0795 | No | ||
| 4 | SEPT9 | 1940040 | 3014 | 0.643 | -0.0922 | No | ||
| 5 | PKN2 | 940746 2350168 | 3193 | 0.577 | -0.0945 | No | ||
| 6 | SSBP2 | 2260725 2900400 4730711 | 3630 | 0.435 | -0.1126 | No | ||
| 7 | CCND1 | 460524 770309 3120576 6980398 | 3689 | 0.419 | -0.1104 | No | ||
| 8 | CDYL | 4200142 4730397 6060400 | 3724 | 0.411 | -0.1070 | No | ||
| 9 | CBLB | 3520465 4670348 | 3829 | 0.390 | -0.1077 | No | ||
| 10 | MGLL | 2030446 4210520 | 3832 | 0.389 | -0.1028 | No | ||
| 11 | IL7R | 2060088 6380500 | 3854 | 0.383 | -0.0991 | No | ||
| 12 | GTF2F2 | 2690546 | 3876 | 0.378 | -0.0954 | No | ||
| 13 | EGFR | 4920138 6480521 | 4005 | 0.351 | -0.0979 | No | ||
| 14 | PPAP2B | 3190397 4730280 | 4078 | 0.339 | -0.0975 | No | ||
| 15 | FGF2 | 3450433 | 4167 | 0.323 | -0.0982 | No | ||
| 16 | EXT1 | 4540603 | 4264 | 0.307 | -0.0995 | No | ||
| 17 | XRCC4 | 1690685 6660427 | 4267 | 0.306 | -0.0957 | No | ||
| 18 | SERPINE1 | 4210403 | 4396 | 0.285 | -0.0990 | No | ||
| 19 | MDN1 | 60671 770091 | 4594 | 0.254 | -0.1064 | No | ||
| 20 | PBX3 | 1300424 3710577 6180575 | 4738 | 0.236 | -0.1111 | No | ||
| 21 | GPR176 | 130017 | 4846 | 0.226 | -0.1141 | No | ||
| 22 | CREB5 | 2320368 | 5120 | 0.191 | -0.1264 | No | ||
| 23 | BMP2K | 60541 5570504 | 5451 | 0.163 | -0.1422 | No | ||
| 24 | GULP1 | 2230725 2350039 5570446 | 5540 | 0.156 | -0.1449 | No | ||
| 25 | CXCL12 | 580546 4150750 4570068 | 5956 | 0.127 | -0.1658 | No | ||
| 26 | FTO | 1690477 | 6087 | 0.119 | -0.1713 | No | ||
| 27 | ATRX | 2340039 3610132 4210373 | 6109 | 0.118 | -0.1709 | No | ||
| 28 | PKP4 | 3710400 5890097 | 6121 | 0.117 | -0.1700 | No | ||
| 29 | EPHA4 | 460750 | 6198 | 0.113 | -0.1727 | No | ||
| 30 | CAP2 | 7100315 | 6254 | 0.110 | -0.1743 | No | ||
| 31 | ATXN1 | 5550156 | 6268 | 0.109 | -0.1736 | No | ||
| 32 | BCAR3 | 1660717 | 6388 | 0.102 | -0.1787 | No | ||
| 33 | FARS2 | 5340575 | 6664 | 0.091 | -0.1924 | No | ||
| 34 | NFIB | 460450 | 6690 | 0.089 | -0.1927 | No | ||
| 35 | GALK2 | 2360632 | 6713 | 0.089 | -0.1927 | No | ||
| 36 | MID1 | 4070114 4760458 | 6716 | 0.089 | -0.1917 | No | ||
| 37 | DDX10 | 520746 | 6767 | 0.086 | -0.1933 | No | ||
| 38 | PTPRM | 6370136 | 7093 | 0.075 | -0.2099 | No | ||
| 39 | GNAQ | 430670 4210131 5900736 | 7189 | 0.072 | -0.2142 | No | ||
| 40 | FBXL7 | 6620711 | 7955 | 0.051 | -0.2549 | No | ||
| 41 | CRADD | 6770520 | 8067 | 0.049 | -0.2603 | No | ||
| 42 | RNF13 | 2370021 | 8379 | 0.042 | -0.2766 | No | ||
| 43 | MYO1B | 770372 | 8443 | 0.041 | -0.2795 | No | ||
| 44 | ROR1 | 6220026 | 8461 | 0.040 | -0.2799 | No | ||
| 45 | NRG1 | 1050332 | 8525 | 0.039 | -0.2828 | No | ||
| 46 | RABGAP1L | 3190014 6860088 | 8556 | 0.038 | -0.2839 | No | ||
| 47 | SKAP2 | 6760500 | 8593 | 0.037 | -0.2854 | No | ||
| 48 | SMAD7 | 430377 | 9300 | 0.023 | -0.3233 | No | ||
| 49 | RGS7 | 3390044 | 9319 | 0.023 | -0.3240 | No | ||
| 50 | CTGF | 4540577 | 9479 | 0.020 | -0.3323 | No | ||
| 51 | GBE1 | 2190368 | 9606 | 0.018 | -0.3389 | No | ||
| 52 | SATB2 | 2470181 | 9856 | 0.014 | -0.3522 | No | ||
| 53 | ERC1 | 1580170 2630717 6110762 6840280 | 9933 | 0.012 | -0.3561 | No | ||
| 54 | SAMD4A | 1170717 | 10258 | 0.007 | -0.3736 | No | ||
| 55 | NEK7 | 4540026 | 10604 | 0.001 | -0.3922 | No | ||
| 56 | SHB | 4920494 | 10628 | 0.001 | -0.3935 | No | ||
| 57 | STAG1 | 1190300 1400722 4590100 | 10654 | 0.000 | -0.3948 | No | ||
| 58 | TLE4 | 1450064 1500519 | 11293 | -0.010 | -0.4292 | No | ||
| 59 | CLASP2 | 2510139 | 11667 | -0.018 | -0.4491 | No | ||
| 60 | VEGFC | 5910494 | 11694 | -0.018 | -0.4503 | No | ||
| 61 | EHBP1 | 430348 | 11986 | -0.024 | -0.4657 | No | ||
| 62 | SCHIP1 | 4560152 | 12061 | -0.026 | -0.4694 | No | ||
| 63 | DPYD | 6110095 | 12303 | -0.031 | -0.4820 | No | ||
| 64 | EDG2 | 6290215 | 12557 | -0.038 | -0.4952 | No | ||
| 65 | FGF5 | 5270204 | 12662 | -0.041 | -0.5004 | No | ||
| 66 | TSPAN5 | 6770019 | 12666 | -0.041 | -0.5000 | No | ||
| 67 | PARD3 | 3390324 5390541 | 12872 | -0.047 | -0.5105 | No | ||
| 68 | CDC42BPA | 840671 | 12955 | -0.050 | -0.5143 | No | ||
| 69 | COL5A2 | 780040 | 13100 | -0.054 | -0.5214 | No | ||
| 70 | F3 | 2940180 | 13140 | -0.056 | -0.5228 | No | ||
| 71 | MYST4 | 1400563 2570687 3360458 6840402 | 13280 | -0.061 | -0.5295 | No | ||
| 72 | LARS2 | 7000706 | 13363 | -0.065 | -0.5331 | No | ||
| 73 | SEMA3C | 7000020 | 13391 | -0.066 | -0.5338 | No | ||
| 74 | FYN | 2100468 4760520 4850687 | 13831 | -0.091 | -0.5564 | No | ||
| 75 | SERPINB2 | 70487 | 13867 | -0.093 | -0.5571 | No | ||
| 76 | PTPRK | 1450433 2640142 | 14037 | -0.106 | -0.5649 | No | ||
| 77 | UST | 2100053 | 14174 | -0.119 | -0.5707 | No | ||
| 78 | DUSP1 | 6860121 | 14399 | -0.144 | -0.5810 | No | ||
| 79 | MYO10 | 3830576 5570092 | 14462 | -0.153 | -0.5824 | No | ||
| 80 | SPAG9 | 580497 2510180 4210685 | 14493 | -0.157 | -0.5820 | No | ||
| 81 | STK39 | 50692 | 14763 | -0.207 | -0.5940 | No | ||
| 82 | RGS20 | 4540541 4730086 | 14926 | -0.242 | -0.5996 | Yes | ||
| 83 | TCF7L2 | 2190348 5220113 | 14949 | -0.248 | -0.5977 | Yes | ||
| 84 | ZFYVE9 | 6980288 7100017 | 14979 | -0.254 | -0.5960 | Yes | ||
| 85 | DOK5 | 770113 3450156 | 15003 | -0.261 | -0.5939 | Yes | ||
| 86 | EIF4E | 1580403 70133 6380215 | 15012 | -0.264 | -0.5910 | Yes | ||
| 87 | CASK | 2340215 3290576 6770215 | 15035 | -0.271 | -0.5888 | Yes | ||
| 88 | LASS6 | 5890576 6520138 | 15109 | -0.294 | -0.5890 | Yes | ||
| 89 | ROCK2 | 1780035 5720025 | 15118 | -0.295 | -0.5857 | Yes | ||
| 90 | STK3 | 7100427 | 15154 | -0.306 | -0.5837 | Yes | ||
| 91 | MYC | 380541 4670170 | 15244 | -0.337 | -0.5842 | Yes | ||
| 92 | NRIP1 | 3190039 | 15347 | -0.369 | -0.5850 | Yes | ||
| 93 | MAD1L1 | 5700035 | 15378 | -0.379 | -0.5819 | Yes | ||
| 94 | CTBP2 | 430309 3710079 | 15569 | -0.445 | -0.5865 | Yes | ||
| 95 | ZNF148 | 6420403 | 15604 | -0.459 | -0.5825 | Yes | ||
| 96 | FUT8 | 1340068 2340056 | 15634 | -0.470 | -0.5781 | Yes | ||
| 97 | AMPH | 2260338 3140576 | 15669 | -0.480 | -0.5738 | Yes | ||
| 98 | MAST2 | 6200348 | 15711 | -0.496 | -0.5697 | Yes | ||
| 99 | FLNB | 2510575 | 15755 | -0.516 | -0.5655 | Yes | ||
| 100 | IFRD1 | 4590215 | 15760 | -0.519 | -0.5591 | Yes | ||
| 101 | THRAP2 | 4920600 | 15773 | -0.524 | -0.5531 | Yes | ||
| 102 | MPHOSPH1 | 4610427 | 15799 | -0.532 | -0.5477 | Yes | ||
| 103 | MAP4K5 | 3190286 4760300 5270070 5550494 | 15930 | -0.591 | -0.5472 | Yes | ||
| 104 | SMURF2 | 1940161 | 15992 | -0.622 | -0.5426 | Yes | ||
| 105 | TDRD3 | 540292 | 16172 | -0.710 | -0.5433 | Yes | ||
| 106 | PPP1R12A | 4070066 4590162 | 16223 | -0.748 | -0.5365 | Yes | ||
| 107 | RNGTT | 780152 780746 1090471 | 16226 | -0.749 | -0.5271 | Yes | ||
| 108 | WDR7 | 670279 1090121 | 16267 | -0.765 | -0.5196 | Yes | ||
| 109 | SLC25A13 | 4920072 5690722 | 16566 | -0.939 | -0.5238 | Yes | ||
| 110 | PRIM2A | 7000692 | 16825 | -1.081 | -0.5240 | Yes | ||
| 111 | MBNL1 | 2640762 7100048 | 17040 | -1.263 | -0.5195 | Yes | ||
| 112 | TCF12 | 3610324 7000156 | 17340 | -1.578 | -0.5156 | Yes | ||
| 113 | CENTG2 | 4210053 | 17380 | -1.626 | -0.4971 | Yes | ||
| 114 | ENC1 | 4810348 | 17562 | -1.828 | -0.4837 | Yes | ||
| 115 | RUNX1 | 3840711 | 17573 | -1.837 | -0.4609 | Yes | ||
| 116 | TBL1X | 6400524 | 17890 | -2.280 | -0.4490 | Yes | ||
| 117 | UBL3 | 6450458 | 17992 | -2.441 | -0.4235 | Yes | ||
| 118 | GSK3B | 5360348 | 18106 | -2.696 | -0.3953 | Yes | ||
| 119 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18147 | -2.771 | -0.3623 | Yes | ||
| 120 | SLC4A7 | 2370056 | 18169 | -2.822 | -0.3276 | Yes | ||
| 121 | FCHSD2 | 5720092 | 18323 | -3.256 | -0.2945 | Yes | ||
| 122 | MNAT1 | 730292 2350364 | 18373 | -3.445 | -0.2534 | Yes | ||
| 123 | EIF4G3 | 670707 | 18431 | -3.687 | -0.2097 | Yes | ||
| 124 | SRPK2 | 6380341 | 18529 | -4.282 | -0.1605 | Yes | ||
| 125 | PRKACB | 4210170 | 18607 | -6.062 | -0.0877 | Yes | ||
| 126 | PTK2 | 1780148 | 18615 | -6.937 | 0.0001 | Yes |