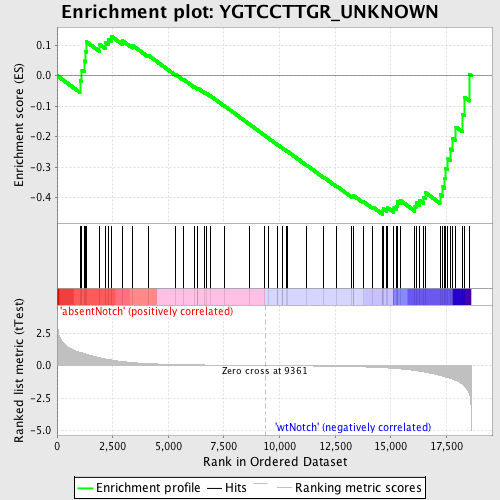
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | YGTCCTTGR_UNKNOWN |
| Enrichment Score (ES) | -0.45580703 |
| Normalized Enrichment Score (NES) | -1.2267659 |
| Nominal p-value | 0.13033175 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC38A2 | 1470242 3800026 | 1029 | 1.006 | -0.0161 | No | ||
| 2 | GAS6 | 4480021 | 1116 | 0.979 | 0.0177 | No | ||
| 3 | MCAM | 5130270 6620577 | 1223 | 0.917 | 0.0479 | No | ||
| 4 | H2AFZ | 1470168 | 1287 | 0.879 | 0.0790 | No | ||
| 5 | HMGCS1 | 6620452 | 1321 | 0.860 | 0.1109 | No | ||
| 6 | HIST1H1A | 1450131 | 1913 | 0.600 | 0.1026 | No | ||
| 7 | CITED2 | 5670114 5130088 | 2190 | 0.502 | 0.1074 | No | ||
| 8 | CS | 5080600 | 2305 | 0.469 | 0.1196 | No | ||
| 9 | THRA | 6770341 110164 1990600 | 2433 | 0.427 | 0.1295 | No | ||
| 10 | ATP5B | 3870138 | 2927 | 0.308 | 0.1150 | No | ||
| 11 | SYNPO2L | 5390719 | 3384 | 0.228 | 0.0994 | No | ||
| 12 | PRKACA | 2640731 4050048 | 4093 | 0.149 | 0.0671 | No | ||
| 13 | GABRA3 | 4810142 | 5315 | 0.080 | 0.0044 | No | ||
| 14 | ZIC3 | 380020 | 5692 | 0.068 | -0.0132 | No | ||
| 15 | PRKCE | 5700053 | 6158 | 0.054 | -0.0362 | No | ||
| 16 | CHST8 | 1940050 | 6317 | 0.051 | -0.0427 | No | ||
| 17 | SEMA6D | 4050324 5860138 6350307 | 6321 | 0.051 | -0.0409 | No | ||
| 18 | MT3 | 1450537 | 6631 | 0.043 | -0.0558 | No | ||
| 19 | DRD3 | 4780402 | 6719 | 0.041 | -0.0589 | No | ||
| 20 | HOXD11 | 2260242 | 6881 | 0.038 | -0.0661 | No | ||
| 21 | KCNJ16 | 4780273 | 7519 | 0.027 | -0.0993 | No | ||
| 22 | SNCB | 3360133 | 8630 | 0.010 | -0.1588 | No | ||
| 23 | PAM | 5290528 | 9329 | 0.000 | -0.1964 | No | ||
| 24 | MLLT6 | 3870168 | 9490 | -0.002 | -0.2049 | No | ||
| 25 | TRIM2 | 430546 1170286 | 9884 | -0.007 | -0.2259 | No | ||
| 26 | EMX2 | 1660092 | 9926 | -0.008 | -0.2278 | No | ||
| 27 | ELAVL2 | 360181 | 10145 | -0.011 | -0.2391 | No | ||
| 28 | AMY2A | 580138 | 10149 | -0.011 | -0.2388 | No | ||
| 29 | SYT7 | 2450528 3390500 3610687 | 10288 | -0.013 | -0.2457 | No | ||
| 30 | NPTX2 | 70021 | 10339 | -0.014 | -0.2479 | No | ||
| 31 | ARPC5 | 1740411 3450300 | 11214 | -0.027 | -0.2939 | No | ||
| 32 | CBX4 | 70332 | 11982 | -0.043 | -0.3336 | No | ||
| 33 | CLCNKA | 1740050 | 12576 | -0.057 | -0.3634 | No | ||
| 34 | PHYHIPL | 2360706 3840692 | 13243 | -0.079 | -0.3962 | No | ||
| 35 | HRH3 | 6180040 | 13314 | -0.082 | -0.3967 | No | ||
| 36 | AFP | 6590358 | 13334 | -0.083 | -0.3945 | No | ||
| 37 | SOCS5 | 3830398 7100093 | 13768 | -0.103 | -0.4138 | No | ||
| 38 | PRKAG1 | 6450091 | 14196 | -0.128 | -0.4318 | No | ||
| 39 | TMEM16D | 2650324 | 14643 | -0.164 | -0.4494 | Yes | ||
| 40 | TNKS1BP1 | 7040184 | 14646 | -0.164 | -0.4430 | Yes | ||
| 41 | FHL2 | 1400008 | 14651 | -0.165 | -0.4368 | Yes | ||
| 42 | ARMET | 5130670 | 14800 | -0.178 | -0.4378 | Yes | ||
| 43 | PANK3 | 3060687 | 14840 | -0.182 | -0.4328 | Yes | ||
| 44 | ATP2A2 | 1090075 3990279 | 15131 | -0.214 | -0.4400 | Yes | ||
| 45 | CYCS | 2030072 | 15141 | -0.216 | -0.4320 | Yes | ||
| 46 | DIRC2 | 2810494 | 15254 | -0.230 | -0.4291 | Yes | ||
| 47 | RGL1 | 1500097 6900450 | 15294 | -0.235 | -0.4219 | Yes | ||
| 48 | SNTA1 | 4570400 | 15312 | -0.238 | -0.4135 | Yes | ||
| 49 | CRAT | 540020 2060364 3520148 | 15435 | -0.256 | -0.4101 | Yes | ||
| 50 | STIM1 | 6380138 | 16084 | -0.379 | -0.4301 | Yes | ||
| 51 | CENTD1 | 2570592 | 16134 | -0.391 | -0.4175 | Yes | ||
| 52 | ESM1 | 1410594 | 16287 | -0.434 | -0.4086 | Yes | ||
| 53 | ADAM11 | 1050008 3130494 | 16476 | -0.486 | -0.3997 | Yes | ||
| 54 | SYT13 | 5910110 | 16560 | -0.513 | -0.3841 | Yes | ||
| 55 | CTRL | 2120301 | 17212 | -0.753 | -0.3897 | Yes | ||
| 56 | YWHAG | 3780341 | 17341 | -0.804 | -0.3651 | Yes | ||
| 57 | HSPBAP1 | 4060022 | 17428 | -0.849 | -0.3364 | Yes | ||
| 58 | PPP2R4 | 670280 2900097 | 17458 | -0.868 | -0.3040 | Yes | ||
| 59 | HDAC3 | 4060072 | 17567 | -0.927 | -0.2735 | Yes | ||
| 60 | OTX2 | 1190471 | 17684 | -0.992 | -0.2408 | Yes | ||
| 61 | BAZ2A | 730184 | 17764 | -1.024 | -0.2050 | Yes | ||
| 62 | VAMP2 | 2100100 | 17926 | -1.144 | -0.1688 | Yes | ||
| 63 | IGSF3 | 6450524 | 18220 | -1.447 | -0.1279 | Yes | ||
| 64 | KCNAB3 | 2470725 | 18303 | -1.577 | -0.0705 | Yes | ||
| 65 | ALDOA | 6290672 | 18544 | -2.228 | 0.0039 | Yes |

