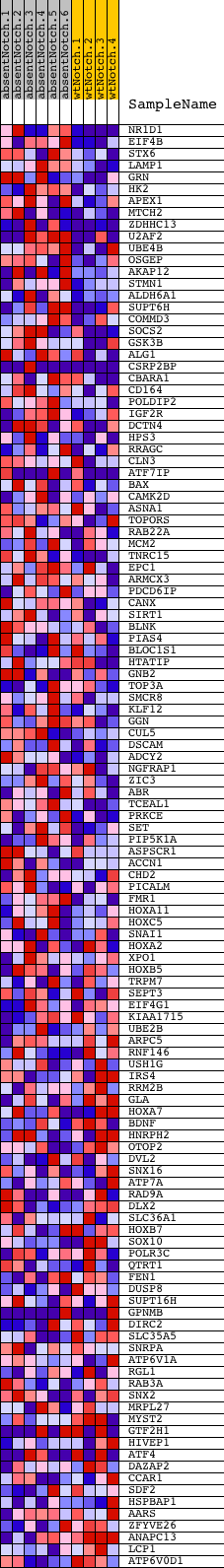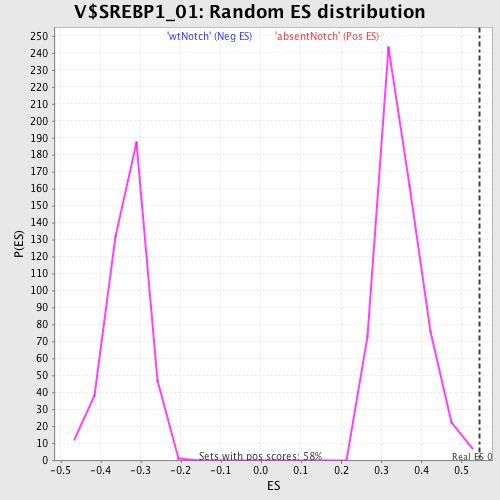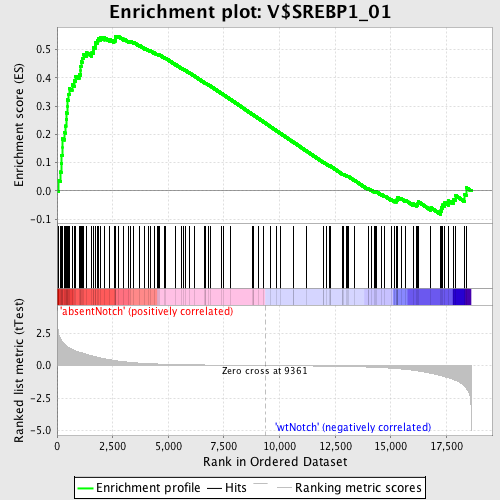
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$SREBP1_01 |
| Enrichment Score (ES) | 0.54709315 |
| Normalized Enrichment Score (NES) | 1.5705445 |
| Nominal p-value | 0.0017152659 |
| FDR q-value | 0.17526667 |
| FWER p-Value | 0.621 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NR1D1 | 2360471 770746 6590204 | 54 | 2.528 | 0.0381 | Yes | ||
| 2 | EIF4B | 5390563 5270577 | 143 | 2.090 | 0.0672 | Yes | ||
| 3 | STX6 | 4850750 6770278 | 182 | 1.963 | 0.0969 | Yes | ||
| 4 | LAMP1 | 2470524 | 203 | 1.920 | 0.1270 | Yes | ||
| 5 | GRN | 4050239 | 236 | 1.839 | 0.1550 | Yes | ||
| 6 | HK2 | 2640722 | 239 | 1.825 | 0.1845 | Yes | ||
| 7 | APEX1 | 3190519 | 326 | 1.637 | 0.2064 | Yes | ||
| 8 | MTCH2 | 3780603 6620215 | 368 | 1.571 | 0.2296 | Yes | ||
| 9 | ZDHHC13 | 430170 4230021 | 400 | 1.538 | 0.2529 | Yes | ||
| 10 | U2AF2 | 450600 4670128 5420292 | 420 | 1.513 | 0.2764 | Yes | ||
| 11 | UBE4B | 780008 3610154 | 453 | 1.473 | 0.2985 | Yes | ||
| 12 | OSGEP | 5700239 | 458 | 1.465 | 0.3221 | Yes | ||
| 13 | AKAP12 | 1450739 | 522 | 1.383 | 0.3411 | Yes | ||
| 14 | STMN1 | 1990717 | 548 | 1.364 | 0.3618 | Yes | ||
| 15 | ALDH6A1 | 1770746 | 673 | 1.249 | 0.3754 | Yes | ||
| 16 | SUPT6H | 3170082 | 773 | 1.182 | 0.3892 | Yes | ||
| 17 | COMMD3 | 2510259 | 828 | 1.142 | 0.4048 | Yes | ||
| 18 | SOCS2 | 4760692 | 988 | 1.024 | 0.4128 | Yes | ||
| 19 | GSK3B | 5360348 | 1055 | 1.000 | 0.4254 | Yes | ||
| 20 | ALG1 | 2570112 | 1059 | 0.999 | 0.4414 | Yes | ||
| 21 | CSRP2BP | 4570717 5550333 | 1082 | 0.996 | 0.4564 | Yes | ||
| 22 | CBARA1 | 360471 | 1127 | 0.973 | 0.4698 | Yes | ||
| 23 | CD164 | 3830594 | 1175 | 0.949 | 0.4826 | Yes | ||
| 24 | POLDIP2 | 2650139 | 1313 | 0.864 | 0.4892 | Yes | ||
| 25 | IGF2R | 1570402 | 1564 | 0.749 | 0.4878 | Yes | ||
| 26 | DCTN4 | 3390010 5550170 | 1627 | 0.716 | 0.4961 | Yes | ||
| 27 | HPS3 | 6650348 | 1635 | 0.711 | 0.5072 | Yes | ||
| 28 | RRAGC | 5910348 | 1711 | 0.681 | 0.5142 | Yes | ||
| 29 | CLN3 | 780368 | 1720 | 0.678 | 0.5248 | Yes | ||
| 30 | ATF7IP | 2690176 6770021 | 1803 | 0.642 | 0.5307 | Yes | ||
| 31 | BAX | 3830008 | 1838 | 0.628 | 0.5391 | Yes | ||
| 32 | CAMK2D | 2370685 | 1962 | 0.579 | 0.5418 | Yes | ||
| 33 | ASNA1 | 3830041 | 2121 | 0.524 | 0.5418 | Yes | ||
| 34 | TOPORS | 4060592 | 2373 | 0.443 | 0.5354 | Yes | ||
| 35 | RAB22A | 110500 3830707 | 2558 | 0.397 | 0.5319 | Yes | ||
| 36 | MCM2 | 5050139 | 2607 | 0.385 | 0.5355 | Yes | ||
| 37 | TNRC15 | 6760746 | 2616 | 0.382 | 0.5413 | Yes | ||
| 38 | EPC1 | 1570050 5290095 6900193 | 2623 | 0.380 | 0.5471 | Yes | ||
| 39 | ARMCX3 | 3290309 | 2743 | 0.349 | 0.5463 | No | ||
| 40 | PDCD6IP | 1990403 | 3003 | 0.293 | 0.5371 | No | ||
| 41 | CANX | 4760402 | 3220 | 0.252 | 0.5295 | No | ||
| 42 | SIRT1 | 1190731 | 3302 | 0.239 | 0.5290 | No | ||
| 43 | BLNK | 70288 6590092 | 3432 | 0.222 | 0.5256 | No | ||
| 44 | PIAS4 | 6040113 | 3710 | 0.187 | 0.5136 | No | ||
| 45 | BLOC1S1 | 130341 3940717 | 3948 | 0.163 | 0.5035 | No | ||
| 46 | HTATIP | 5360739 | 4094 | 0.149 | 0.4980 | No | ||
| 47 | GNB2 | 2350053 | 4180 | 0.142 | 0.4958 | No | ||
| 48 | TOP3A | 7000435 | 4362 | 0.129 | 0.4881 | No | ||
| 49 | SMCR8 | 730653 | 4499 | 0.120 | 0.4827 | No | ||
| 50 | KLF12 | 1660095 4810288 5340546 6520286 | 4562 | 0.117 | 0.4812 | No | ||
| 51 | GGN | 1410528 4210082 5290170 | 4605 | 0.113 | 0.4808 | No | ||
| 52 | CUL5 | 450142 | 4818 | 0.101 | 0.4709 | No | ||
| 53 | DSCAM | 1780050 2450731 2810438 | 4891 | 0.098 | 0.4686 | No | ||
| 54 | ADCY2 | 4610100 | 5334 | 0.080 | 0.4460 | No | ||
| 55 | NGFRAP1 | 6770195 | 5585 | 0.071 | 0.4337 | No | ||
| 56 | ZIC3 | 380020 | 5692 | 0.068 | 0.4290 | No | ||
| 57 | ABR | 610079 1170609 3610195 5670050 | 5777 | 0.065 | 0.4255 | No | ||
| 58 | TCEAL1 | 870577 | 5965 | 0.059 | 0.4164 | No | ||
| 59 | PRKCE | 5700053 | 6158 | 0.054 | 0.4069 | No | ||
| 60 | SET | 6650286 | 6630 | 0.043 | 0.3821 | No | ||
| 61 | PIP5K1A | 4560672 | 6681 | 0.042 | 0.3801 | No | ||
| 62 | ASPSCR1 | 610167 3990739 | 6689 | 0.042 | 0.3804 | No | ||
| 63 | ACCN1 | 2060139 2190541 | 6823 | 0.040 | 0.3739 | No | ||
| 64 | CHD2 | 4070039 | 6889 | 0.038 | 0.3710 | No | ||
| 65 | PICALM | 1940280 | 7366 | 0.030 | 0.3457 | No | ||
| 66 | FMR1 | 5050075 | 7485 | 0.028 | 0.3398 | No | ||
| 67 | HOXA11 | 6980133 | 7781 | 0.023 | 0.3242 | No | ||
| 68 | HOXC5 | 1580017 | 8766 | 0.008 | 0.2711 | No | ||
| 69 | SNAI1 | 6590253 | 8820 | 0.007 | 0.2684 | No | ||
| 70 | HOXA2 | 2120121 | 8839 | 0.007 | 0.2675 | No | ||
| 71 | XPO1 | 540707 | 9072 | 0.004 | 0.2550 | No | ||
| 72 | HOXB5 | 2900538 | 9272 | 0.001 | 0.2443 | No | ||
| 73 | TRPM7 | 2510408 | 9576 | -0.003 | 0.2280 | No | ||
| 74 | SEPT3 | 3830022 5290079 | 9845 | -0.006 | 0.2136 | No | ||
| 75 | EIF4G1 | 4070446 | 10031 | -0.009 | 0.2037 | No | ||
| 76 | KIAA1715 | 1190551 | 10602 | -0.018 | 0.1732 | No | ||
| 77 | UBE2B | 4780497 | 10616 | -0.018 | 0.1728 | No | ||
| 78 | ARPC5 | 1740411 3450300 | 11214 | -0.027 | 0.1409 | No | ||
| 79 | RNF146 | 1660735 | 11952 | -0.042 | 0.1018 | No | ||
| 80 | USH1G | 2940446 | 11986 | -0.043 | 0.1007 | No | ||
| 81 | IRS4 | 430341 1190450 | 12103 | -0.045 | 0.0951 | No | ||
| 82 | RRM2B | 1690025 1690465 4010400 | 12247 | -0.048 | 0.0882 | No | ||
| 83 | GLA | 4610364 | 12268 | -0.049 | 0.0879 | No | ||
| 84 | HOXA7 | 5910152 | 12270 | -0.049 | 0.0886 | No | ||
| 85 | BDNF | 2940128 3520368 | 12822 | -0.064 | 0.0599 | No | ||
| 86 | HNRPH2 | 2630040 5720671 | 12847 | -0.065 | 0.0596 | No | ||
| 87 | OTOP2 | 610068 | 12886 | -0.066 | 0.0586 | No | ||
| 88 | DVL2 | 6110162 | 12999 | -0.070 | 0.0537 | No | ||
| 89 | SNX16 | 870446 | 13045 | -0.072 | 0.0525 | No | ||
| 90 | ATP7A | 6550168 | 13085 | -0.073 | 0.0515 | No | ||
| 91 | RAD9A | 110300 1940632 3990390 6040014 | 13348 | -0.084 | 0.0387 | No | ||
| 92 | DLX2 | 2320438 4200673 | 13981 | -0.115 | 0.0064 | No | ||
| 93 | SLC36A1 | 5080242 | 13996 | -0.116 | 0.0075 | No | ||
| 94 | HOXB7 | 4540706 4610358 | 14123 | -0.123 | 0.0027 | No | ||
| 95 | SOX10 | 6200538 | 14283 | -0.134 | -0.0037 | No | ||
| 96 | POLR3C | 6940546 | 14311 | -0.136 | -0.0030 | No | ||
| 97 | QTRT1 | 1340162 | 14349 | -0.139 | -0.0027 | No | ||
| 98 | FEN1 | 1770541 | 14598 | -0.160 | -0.0135 | No | ||
| 99 | DUSP8 | 6650039 | 14730 | -0.171 | -0.0178 | No | ||
| 100 | SUPT16H | 1240333 | 15022 | -0.200 | -0.0303 | No | ||
| 101 | GPNMB | 1780138 2340176 3390403 4560156 | 15185 | -0.221 | -0.0355 | No | ||
| 102 | DIRC2 | 2810494 | 15254 | -0.230 | -0.0354 | No | ||
| 103 | SLC35A5 | 2970670 4480546 | 15258 | -0.231 | -0.0319 | No | ||
| 104 | SNRPA | 2570100 4150121 6940484 | 15287 | -0.234 | -0.0296 | No | ||
| 105 | ATP6V1A | 6590242 | 15291 | -0.234 | -0.0259 | No | ||
| 106 | RGL1 | 1500097 6900450 | 15294 | -0.235 | -0.0222 | No | ||
| 107 | RAB3A | 3940288 | 15458 | -0.259 | -0.0268 | No | ||
| 108 | SNX2 | 2470068 3360364 6510064 | 15672 | -0.298 | -0.0335 | No | ||
| 109 | MRPL27 | 2060131 | 16007 | -0.363 | -0.0457 | No | ||
| 110 | MYST2 | 4540494 | 16154 | -0.398 | -0.0472 | No | ||
| 111 | GTF2H1 | 2570746 2650148 | 16213 | -0.412 | -0.0436 | No | ||
| 112 | HIVEP1 | 3850632 | 16240 | -0.420 | -0.0382 | No | ||
| 113 | ATF4 | 730441 1740195 | 16803 | -0.593 | -0.0590 | No | ||
| 114 | DAZAP2 | 6200102 | 17250 | -0.770 | -0.0706 | No | ||
| 115 | CCAR1 | 3190092 | 17272 | -0.777 | -0.0592 | No | ||
| 116 | SDF2 | 2690601 | 17332 | -0.800 | -0.0494 | No | ||
| 117 | HSPBAP1 | 4060022 | 17428 | -0.849 | -0.0408 | No | ||
| 118 | AARS | 1570279 2370136 | 17602 | -0.940 | -0.0349 | No | ||
| 119 | ZFYVE26 | 6860152 | 17806 | -1.060 | -0.0287 | No | ||
| 120 | ANAPC13 | 3800446 | 17910 | -1.133 | -0.0159 | No | ||
| 121 | LCP1 | 4280397 | 18306 | -1.578 | -0.0117 | No | ||
| 122 | ATP6V0D1 | 360348 5900021 6290605 | 18398 | -1.752 | 0.0118 | No |