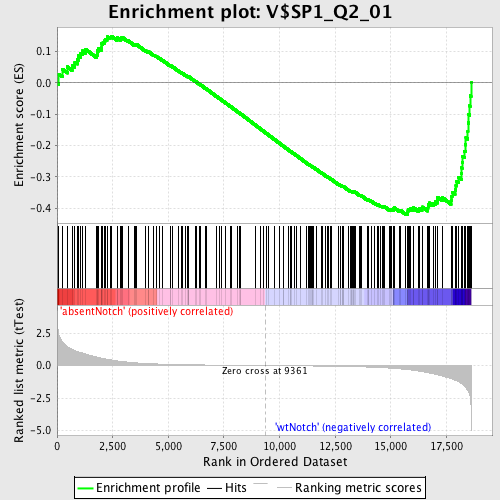
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$SP1_Q2_01 |
| Enrichment Score (ES) | -0.42005542 |
| Normalized Enrichment Score (NES) | -1.3236885 |
| Nominal p-value | 0.012376238 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PER1 | 3290315 | 76 | 2.373 | 0.0280 | No | ||
| 2 | KLF7 | 50390 | 241 | 1.820 | 0.0437 | No | ||
| 3 | BCL11B | 2680673 | 462 | 1.464 | 0.0516 | No | ||
| 4 | ZFP36 | 2030605 | 675 | 1.248 | 0.0570 | No | ||
| 5 | ACLY | 1740609 | 799 | 1.163 | 0.0660 | No | ||
| 6 | RAP1GDS1 | 50528 | 902 | 1.086 | 0.0752 | No | ||
| 7 | ETV5 | 110017 | 949 | 1.049 | 0.0869 | No | ||
| 8 | FAF1 | 2450717 2510121 4730156 | 1047 | 1.001 | 0.0952 | No | ||
| 9 | GRB2 | 6650398 | 1153 | 0.960 | 0.1025 | No | ||
| 10 | SNAG1 | 2760092 | 1288 | 0.879 | 0.1071 | No | ||
| 11 | BCL6 | 940100 | 1780 | 0.649 | 0.0893 | No | ||
| 12 | TPM4 | 2100059 3850072 4920162 | 1802 | 0.642 | 0.0968 | No | ||
| 13 | ANP32A | 510044 1580095 2680037 | 1825 | 0.634 | 0.1042 | No | ||
| 14 | PIP5K2B | 6520156 6620541 | 1870 | 0.617 | 0.1101 | No | ||
| 15 | BMI1 | 3830026 2630204 | 1980 | 0.573 | 0.1120 | No | ||
| 16 | LASP1 | 1690528 2190341 6420341 | 1995 | 0.569 | 0.1189 | No | ||
| 17 | HIPK1 | 110193 | 1996 | 0.569 | 0.1266 | No | ||
| 18 | HS2ST1 | 3170088 | 2061 | 0.547 | 0.1306 | No | ||
| 19 | ARFIP1 | 3130601 | 2123 | 0.524 | 0.1343 | No | ||
| 20 | LDB1 | 5270601 | 2154 | 0.512 | 0.1396 | No | ||
| 21 | NFE2L1 | 6130053 | 2244 | 0.489 | 0.1414 | No | ||
| 22 | TCF12 | 3610324 7000156 | 2257 | 0.484 | 0.1473 | No | ||
| 23 | ARPC4 | 2900056 | 2382 | 0.441 | 0.1466 | No | ||
| 24 | THRA | 6770341 110164 1990600 | 2433 | 0.427 | 0.1496 | No | ||
| 25 | HYAL2 | 1500152 | 2695 | 0.361 | 0.1404 | No | ||
| 26 | GABARAP | 1450286 | 2700 | 0.360 | 0.1450 | No | ||
| 27 | SH3BP5 | 7100408 | 2869 | 0.322 | 0.1403 | No | ||
| 28 | FUT8 | 1340068 2340056 | 2878 | 0.320 | 0.1441 | No | ||
| 29 | CLPTM1 | 670181 | 2932 | 0.307 | 0.1454 | No | ||
| 30 | KCNN2 | 990025 3520524 | 3192 | 0.258 | 0.1349 | No | ||
| 31 | TBX3 | 2570672 | 3476 | 0.214 | 0.1224 | No | ||
| 32 | CLSTN1 | 6020019 | 3511 | 0.210 | 0.1234 | No | ||
| 33 | SCARF2 | 5290184 | 3579 | 0.202 | 0.1225 | No | ||
| 34 | BCL7C | 460050 | 3994 | 0.159 | 0.1022 | No | ||
| 35 | KLF16 | 4590725 | 4100 | 0.149 | 0.0985 | No | ||
| 36 | ASCL2 | 1400273 | 4104 | 0.148 | 0.1004 | No | ||
| 37 | PIAS3 | 50204 240739 4060524 | 4345 | 0.131 | 0.0891 | No | ||
| 38 | RNF24 | 4230280 6100070 6620739 | 4456 | 0.123 | 0.0848 | No | ||
| 39 | GGN | 1410528 4210082 5290170 | 4605 | 0.113 | 0.0783 | No | ||
| 40 | CAMK2G | 630097 | 4725 | 0.106 | 0.0733 | No | ||
| 41 | HSD11B2 | 5900053 | 5076 | 0.090 | 0.0556 | No | ||
| 42 | DUSP15 | 1850044 6960196 | 5106 | 0.089 | 0.0552 | No | ||
| 43 | GPR3 | 2970452 | 5205 | 0.085 | 0.0510 | No | ||
| 44 | BCL6B | 60047 | 5469 | 0.075 | 0.0378 | No | ||
| 45 | EDG8 | 1990441 | 5595 | 0.071 | 0.0320 | No | ||
| 46 | LOXL4 | 6520095 | 5618 | 0.070 | 0.0317 | No | ||
| 47 | CDH3 | 5390112 6860603 | 5756 | 0.066 | 0.0252 | No | ||
| 48 | KLF13 | 4670563 | 5870 | 0.062 | 0.0199 | No | ||
| 49 | TSNAXIP1 | 1770270 6180577 | 5890 | 0.062 | 0.0197 | No | ||
| 50 | APLP2 | 3610037 | 5901 | 0.061 | 0.0200 | No | ||
| 51 | PAK1 | 4540315 | 5902 | 0.061 | 0.0208 | No | ||
| 52 | NXPH4 | 3830500 | 6211 | 0.053 | 0.0048 | No | ||
| 53 | PTCH2 | 2900092 | 6245 | 0.053 | 0.0037 | No | ||
| 54 | SP3 | 3840338 | 6387 | 0.049 | -0.0032 | No | ||
| 55 | FGF5 | 5270204 | 6443 | 0.047 | -0.0056 | No | ||
| 56 | KCNB1 | 3610035 | 6691 | 0.042 | -0.0184 | No | ||
| 57 | JUB | 5860711 5900500 | 6711 | 0.042 | -0.0189 | No | ||
| 58 | SOX2 | 630487 2230491 | 7148 | 0.033 | -0.0421 | No | ||
| 59 | ACCN2 | 6450465 | 7301 | 0.031 | -0.0499 | No | ||
| 60 | TIGD4 | 1780114 | 7396 | 0.029 | -0.0546 | No | ||
| 61 | KCNMA1 | 1450402 | 7589 | 0.026 | -0.0647 | No | ||
| 62 | NLK | 2030010 2450041 | 7808 | 0.022 | -0.0762 | No | ||
| 63 | ERBB3 | 5890372 | 7817 | 0.022 | -0.0763 | No | ||
| 64 | ADRB1 | 6860292 | 8090 | 0.018 | -0.0908 | No | ||
| 65 | NUMBL | 6130360 6200333 | 8195 | 0.016 | -0.0963 | No | ||
| 66 | KCNIP2 | 60088 1780324 | 8257 | 0.015 | -0.0994 | No | ||
| 67 | ADAM10 | 3780156 | 8259 | 0.015 | -0.0992 | No | ||
| 68 | DNMT3A | 4050068 4810717 | 8894 | 0.006 | -0.1335 | No | ||
| 69 | NXPH3 | 5860292 | 9152 | 0.003 | -0.1474 | No | ||
| 70 | DES | 1450341 | 9284 | 0.001 | -0.1545 | No | ||
| 71 | GRIN2A | 6550538 | 9404 | -0.001 | -0.1609 | No | ||
| 72 | CACNA1A | 6220164 | 9520 | -0.002 | -0.1671 | No | ||
| 73 | NOL4 | 5050446 5360519 | 9748 | -0.005 | -0.1794 | No | ||
| 74 | CRY1 | 6770039 | 9973 | -0.008 | -0.1914 | No | ||
| 75 | PTHLH | 5290739 | 10160 | -0.011 | -0.2014 | No | ||
| 76 | FEV | 840369 5860139 | 10172 | -0.011 | -0.2018 | No | ||
| 77 | AKT2 | 3850541 3870519 | 10409 | -0.015 | -0.2144 | No | ||
| 78 | PTPN2 | 130315 | 10503 | -0.016 | -0.2192 | No | ||
| 79 | OSBPL9 | 2570373 | 10511 | -0.017 | -0.2194 | No | ||
| 80 | RAB26 | 5910025 | 10519 | -0.017 | -0.2195 | No | ||
| 81 | PTPRJ | 4010707 | 10676 | -0.019 | -0.2277 | No | ||
| 82 | BHLHB3 | 3450438 | 10774 | -0.020 | -0.2327 | No | ||
| 83 | CHAT | 6840603 | 10949 | -0.023 | -0.2419 | No | ||
| 84 | NTF5 | 6110239 | 11201 | -0.027 | -0.2551 | No | ||
| 85 | CLDN11 | 670338 | 11280 | -0.029 | -0.2589 | No | ||
| 86 | FSTL5 | 770280 | 11336 | -0.030 | -0.2615 | No | ||
| 87 | TIMELESS | 3710315 | 11391 | -0.031 | -0.2640 | No | ||
| 88 | GSC | 1580685 | 11436 | -0.031 | -0.2660 | No | ||
| 89 | PHOX2A | 6650121 | 11472 | -0.032 | -0.2675 | No | ||
| 90 | NCOA6 | 1780333 6450110 | 11496 | -0.033 | -0.2683 | No | ||
| 91 | OSBP2 | 6510368 | 11536 | -0.034 | -0.2699 | No | ||
| 92 | POLR2I | 4060400 | 11658 | -0.036 | -0.2760 | No | ||
| 93 | MDS1 | 6400072 | 11890 | -0.040 | -0.2880 | No | ||
| 94 | SLC18A3 | 3060035 | 11942 | -0.042 | -0.2902 | No | ||
| 95 | NR2F2 | 3170609 3310577 | 12079 | -0.045 | -0.2970 | No | ||
| 96 | FZD2 | 6480154 | 12152 | -0.046 | -0.3002 | No | ||
| 97 | TADA3L | 4210070 5720088 6660333 | 12206 | -0.047 | -0.3025 | No | ||
| 98 | FGF11 | 840292 | 12271 | -0.049 | -0.3053 | No | ||
| 99 | TBX2 | 1990563 | 12335 | -0.051 | -0.3080 | No | ||
| 100 | RANBP10 | 2340184 | 12660 | -0.059 | -0.3248 | No | ||
| 101 | ONECUT1 | 6450136 | 12728 | -0.061 | -0.3276 | No | ||
| 102 | TITF1 | 3390554 4540292 | 12754 | -0.062 | -0.3281 | No | ||
| 103 | BDNF | 2940128 3520368 | 12822 | -0.064 | -0.3309 | No | ||
| 104 | SIX4 | 6760022 | 12830 | -0.064 | -0.3304 | No | ||
| 105 | HCN4 | 1050026 | 12854 | -0.065 | -0.3308 | No | ||
| 106 | FGF12 | 1740446 2360037 | 12880 | -0.066 | -0.3312 | No | ||
| 107 | EFNB3 | 5570594 | 13115 | -0.075 | -0.3429 | No | ||
| 108 | CDH2 | 520435 2450451 2760025 6650477 | 13177 | -0.077 | -0.3452 | No | ||
| 109 | ENAH | 1690292 5700300 | 13222 | -0.078 | -0.3465 | No | ||
| 110 | SLC35F5 | 2360369 | 13245 | -0.079 | -0.3466 | No | ||
| 111 | CDH24 | 6980592 | 13284 | -0.081 | -0.3476 | No | ||
| 112 | HRH3 | 6180040 | 13314 | -0.082 | -0.3480 | No | ||
| 113 | DLX3 | 3190497 | 13337 | -0.084 | -0.3481 | No | ||
| 114 | ARHGEF19 | 940102 | 13344 | -0.084 | -0.3473 | No | ||
| 115 | CREB1 | 1500717 2230358 3610600 6550601 | 13420 | -0.087 | -0.3502 | No | ||
| 116 | WNT1 | 4780148 | 13613 | -0.095 | -0.3593 | No | ||
| 117 | SLITRK5 | 6590292 | 13614 | -0.095 | -0.3580 | No | ||
| 118 | TAL1 | 7040239 | 13685 | -0.098 | -0.3605 | No | ||
| 119 | TRIM28 | 380292 | 13934 | -0.113 | -0.3724 | No | ||
| 120 | MAP3K11 | 7000039 | 13987 | -0.115 | -0.3737 | No | ||
| 121 | RAB2B | 6200091 | 14012 | -0.116 | -0.3734 | No | ||
| 122 | HOXB7 | 4540706 4610358 | 14123 | -0.123 | -0.3777 | No | ||
| 123 | HCFC1R1 | 6350500 | 14266 | -0.133 | -0.3836 | No | ||
| 124 | CBLN1 | 5670358 | 14392 | -0.142 | -0.3885 | No | ||
| 125 | EN1 | 5360168 7050025 | 14434 | -0.145 | -0.3888 | No | ||
| 126 | EGLN2 | 540086 | 14556 | -0.156 | -0.3932 | No | ||
| 127 | PPRC1 | 1740441 | 14616 | -0.161 | -0.3942 | No | ||
| 128 | ERG | 50154 1770739 | 14665 | -0.166 | -0.3946 | No | ||
| 129 | SPAG9 | 580497 2510180 4210685 | 14719 | -0.170 | -0.3952 | No | ||
| 130 | CACNA1D | 1740315 4730731 | 14960 | -0.194 | -0.4056 | No | ||
| 131 | EIF2C1 | 6900551 | 14965 | -0.194 | -0.4032 | No | ||
| 132 | ACE | 1780161 4920315 | 15041 | -0.203 | -0.4045 | No | ||
| 133 | WNT5A | 840685 3120152 | 15104 | -0.211 | -0.4050 | No | ||
| 134 | ATP2A2 | 1090075 3990279 | 15131 | -0.214 | -0.4035 | No | ||
| 135 | FBXO34 | 4010671 5700647 6450026 | 15133 | -0.215 | -0.4006 | No | ||
| 136 | PHLDA3 | 510398 | 15142 | -0.216 | -0.3982 | No | ||
| 137 | NTRK3 | 4050687 4070167 4610026 6100484 | 15400 | -0.251 | -0.4087 | No | ||
| 138 | ADCK1 | 6620707 | 15424 | -0.254 | -0.4065 | No | ||
| 139 | SNX2 | 2470068 3360364 6510064 | 15672 | -0.298 | -0.4159 | Yes | ||
| 140 | RELB | 1400048 | 15750 | -0.311 | -0.4158 | Yes | ||
| 141 | ASB7 | 520242 2450088 3990563 4280538 | 15762 | -0.313 | -0.4122 | Yes | ||
| 142 | CDK5R1 | 3870161 | 15770 | -0.314 | -0.4083 | Yes | ||
| 143 | NFAT5 | 2510411 5890195 6550152 | 15785 | -0.317 | -0.4048 | Yes | ||
| 144 | MAP4K2 | 4200037 | 15850 | -0.331 | -0.4038 | Yes | ||
| 145 | CHKA | 510324 | 15898 | -0.340 | -0.4017 | Yes | ||
| 146 | PAX2 | 6040270 7000133 | 16006 | -0.363 | -0.4026 | Yes | ||
| 147 | CORO1C | 4570397 | 16012 | -0.364 | -0.3980 | Yes | ||
| 148 | IRF2BP1 | 3170019 | 16258 | -0.426 | -0.4055 | Yes | ||
| 149 | GGA1 | 70402 380168 | 16277 | -0.431 | -0.4007 | Yes | ||
| 150 | CGGBP1 | 4810053 | 16403 | -0.468 | -0.4011 | Yes | ||
| 151 | LYL1 | 2900242 | 16430 | -0.476 | -0.3961 | Yes | ||
| 152 | SESN2 | 2370019 | 16670 | -0.550 | -0.4016 | Yes | ||
| 153 | HAP1 | 1240010 4230079 6980195 | 16672 | -0.551 | -0.3942 | Yes | ||
| 154 | SIPA1 | 5220687 | 16698 | -0.559 | -0.3880 | Yes | ||
| 155 | FBXO3 | 6760673 6860131 | 16753 | -0.575 | -0.3831 | Yes | ||
| 156 | GAS7 | 2120358 6450494 | 16924 | -0.631 | -0.3838 | Yes | ||
| 157 | GFRA1 | 3610152 | 17009 | -0.668 | -0.3793 | Yes | ||
| 158 | RELA | 3830075 | 17087 | -0.699 | -0.3741 | Yes | ||
| 159 | BAD | 2120148 | 17103 | -0.707 | -0.3653 | Yes | ||
| 160 | SMARCD1 | 3060193 3850184 6400369 | 17317 | -0.794 | -0.3661 | Yes | ||
| 161 | MXD4 | 4810113 | 17706 | -0.999 | -0.3737 | Yes | ||
| 162 | TEF | 1050050 3120397 4060487 | 17737 | -1.008 | -0.3616 | Yes | ||
| 163 | CRYAB | 4810619 | 17755 | -1.021 | -0.3488 | Yes | ||
| 164 | DDX6 | 2630025 6770408 | 17886 | -1.117 | -0.3407 | Yes | ||
| 165 | VAMP2 | 2100100 | 17926 | -1.144 | -0.3273 | Yes | ||
| 166 | UPF2 | 4810471 | 17961 | -1.173 | -0.3133 | Yes | ||
| 167 | HSPB2 | 1990110 | 18051 | -1.246 | -0.3013 | Yes | ||
| 168 | RHOG | 6760575 | 18173 | -1.375 | -0.2892 | Yes | ||
| 169 | OAZ2 | 2810110 | 18190 | -1.396 | -0.2712 | Yes | ||
| 170 | FBXL19 | 4570039 | 18205 | -1.420 | -0.2527 | Yes | ||
| 171 | CAMKK1 | 6370324 | 18219 | -1.447 | -0.2339 | Yes | ||
| 172 | BCL3 | 3990440 | 18315 | -1.592 | -0.2175 | Yes | ||
| 173 | SRR | 1190465 3800494 | 18352 | -1.663 | -0.1969 | Yes | ||
| 174 | SEP15 | 1780451 2060450 5890451 | 18376 | -1.724 | -0.1749 | Yes | ||
| 175 | EFEMP2 | 5550086 | 18462 | -1.925 | -0.1534 | Yes | ||
| 176 | TMEM24 | 5860039 | 18469 | -1.952 | -0.1273 | Yes | ||
| 177 | PIK4CB | 510450 1780452 | 18500 | -2.061 | -0.1011 | Yes | ||
| 178 | PAK4 | 1690152 | 18548 | -2.254 | -0.0731 | Yes | ||
| 179 | NOTCH3 | 3060603 | 18565 | -2.482 | -0.0404 | Yes | ||
| 180 | BCL9L | 6940053 | 18607 | -3.187 | 0.0005 | Yes |

