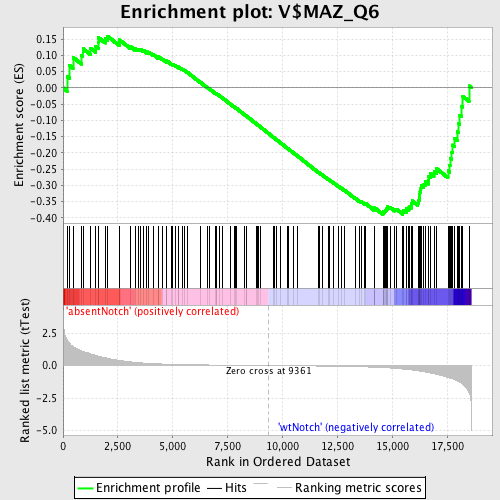
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$MAZ_Q6 |
| Enrichment Score (ES) | -0.38930586 |
| Normalized Enrichment Score (NES) | -1.1651558 |
| Nominal p-value | 0.14628297 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | STX6 | 4850750 6770278 | 182 | 1.963 | 0.0353 | No | ||
| 2 | SYT11 | 630433 | 289 | 1.713 | 0.0690 | No | ||
| 3 | UBE4B | 780008 3610154 | 453 | 1.473 | 0.0941 | No | ||
| 4 | CNN3 | 6110020 | 839 | 1.131 | 0.0993 | No | ||
| 5 | NEURL | 5390538 | 906 | 1.083 | 0.1206 | No | ||
| 6 | SLC2A1 | 2100609 | 1257 | 0.898 | 0.1224 | No | ||
| 7 | FXC1 | 3710053 4060575 | 1489 | 0.779 | 0.1278 | No | ||
| 8 | PLK3 | 2640592 | 1600 | 0.729 | 0.1386 | No | ||
| 9 | LMNA | 520471 1500075 3190167 4210020 | 1609 | 0.724 | 0.1548 | No | ||
| 10 | TCEA3 | 1770020 | 1920 | 0.597 | 0.1518 | No | ||
| 11 | HDAC1 | 2850670 | 2020 | 0.560 | 0.1593 | No | ||
| 12 | JMJD2A | 3610128 4200373 | 2565 | 0.394 | 0.1390 | No | ||
| 13 | NRAS | 6900577 | 2573 | 0.393 | 0.1477 | No | ||
| 14 | EIF4G2 | 3800575 6860184 | 3081 | 0.278 | 0.1266 | No | ||
| 15 | SETDB1 | 840093 | 3295 | 0.240 | 0.1207 | No | ||
| 16 | RPL5 | 840095 | 3416 | 0.224 | 0.1193 | No | ||
| 17 | CLSTN1 | 6020019 | 3511 | 0.210 | 0.1191 | No | ||
| 18 | TCF7L2 | 2190348 5220113 | 3664 | 0.193 | 0.1153 | No | ||
| 19 | TRIM8 | 6130441 | 3820 | 0.173 | 0.1109 | No | ||
| 20 | JUN | 840170 | 3909 | 0.166 | 0.1099 | No | ||
| 21 | ASCL2 | 1400273 | 4104 | 0.148 | 0.1029 | No | ||
| 22 | THRAP3 | 6180309 | 4330 | 0.132 | 0.0937 | No | ||
| 23 | PIAS3 | 50204 240739 4060524 | 4345 | 0.131 | 0.0960 | No | ||
| 24 | PRELP | 5900390 | 4527 | 0.118 | 0.0889 | No | ||
| 25 | NPPA | 6550328 | 4724 | 0.106 | 0.0808 | No | ||
| 26 | CAMK2G | 630097 | 4725 | 0.106 | 0.0832 | No | ||
| 27 | KCNC1 | 770110 | 4955 | 0.095 | 0.0730 | No | ||
| 28 | CUGBP1 | 450292 510022 7050176 7050215 | 5006 | 0.093 | 0.0725 | No | ||
| 29 | PIK3AP1 | 6760008 | 5119 | 0.088 | 0.0684 | No | ||
| 30 | ST5 | 3780204 | 5256 | 0.083 | 0.0630 | No | ||
| 31 | MGEA5 | 6100014 | 5262 | 0.083 | 0.0646 | No | ||
| 32 | CREM | 840156 6380438 6660041 6660168 | 5425 | 0.076 | 0.0576 | No | ||
| 33 | DDX50 | 1450059 2680170 | 5515 | 0.073 | 0.0545 | No | ||
| 34 | HCRTR1 | 1580273 | 5664 | 0.069 | 0.0481 | No | ||
| 35 | CUL2 | 4200278 | 6239 | 0.053 | 0.0182 | No | ||
| 36 | WAC | 5340390 5910164 | 6570 | 0.045 | 0.0014 | No | ||
| 37 | PIP5K1A | 4560672 | 6681 | 0.042 | -0.0036 | No | ||
| 38 | PBX1 | 6660301 | 6950 | 0.037 | -0.0172 | No | ||
| 39 | LHX4 | 6940551 | 6999 | 0.036 | -0.0190 | No | ||
| 40 | SHC1 | 2900731 3170504 6520537 | 7007 | 0.036 | -0.0185 | No | ||
| 41 | HPCA | 6860010 | 7114 | 0.034 | -0.0235 | No | ||
| 42 | RNF2 | 2340497 | 7250 | 0.031 | -0.0300 | No | ||
| 43 | SCUBE2 | 2850315 5130040 | 7638 | 0.025 | -0.0504 | No | ||
| 44 | TLX1 | 2630162 | 7818 | 0.022 | -0.0596 | No | ||
| 45 | SSBP3 | 870717 1050377 7040551 | 7837 | 0.022 | -0.0600 | No | ||
| 46 | PAX6 | 1190025 | 7921 | 0.021 | -0.0640 | No | ||
| 47 | KCNIP2 | 60088 1780324 | 8257 | 0.015 | -0.0818 | No | ||
| 48 | PRDM16 | 4120541 | 8354 | 0.014 | -0.0867 | No | ||
| 49 | HNRPF | 670138 | 8367 | 0.014 | -0.0870 | No | ||
| 50 | EFNA1 | 3840672 | 8792 | 0.008 | -0.1097 | No | ||
| 51 | RTN4RL2 | 6450609 | 8864 | 0.007 | -0.1134 | No | ||
| 52 | POGZ | 870348 7040286 | 8907 | 0.006 | -0.1156 | No | ||
| 53 | LIN28 | 6590672 | 9003 | 0.005 | -0.1206 | No | ||
| 54 | APBB1 | 2690338 | 9605 | -0.003 | -0.1530 | No | ||
| 55 | XPR1 | 1500093 | 9612 | -0.003 | -0.1533 | No | ||
| 56 | PROX1 | 4210193 | 9706 | -0.004 | -0.1582 | No | ||
| 57 | EMX2 | 1660092 | 9926 | -0.008 | -0.1698 | No | ||
| 58 | BCL9 | 7100112 | 10228 | -0.012 | -0.1858 | No | ||
| 59 | SYT7 | 2450528 3390500 3610687 | 10288 | -0.013 | -0.1887 | No | ||
| 60 | KCNQ4 | 3800438 7050400 | 10510 | -0.016 | -0.2003 | No | ||
| 61 | PTPRJ | 4010707 | 10676 | -0.019 | -0.2088 | No | ||
| 62 | GRK5 | 1940348 4670053 | 11641 | -0.036 | -0.2601 | No | ||
| 63 | ROR1 | 6220026 | 11699 | -0.037 | -0.2623 | No | ||
| 64 | ENSA | 6620546 | 11800 | -0.039 | -0.2669 | No | ||
| 65 | NR0B2 | 2470692 | 12101 | -0.045 | -0.2820 | No | ||
| 66 | MYOZ1 | 2760670 | 12149 | -0.046 | -0.2835 | No | ||
| 67 | MYBPC3 | 650390 | 12320 | -0.050 | -0.2916 | No | ||
| 68 | RERE | 3940279 | 12555 | -0.056 | -0.3029 | No | ||
| 69 | NFYC | 1400746 | 12670 | -0.059 | -0.3077 | No | ||
| 70 | BDNF | 2940128 3520368 | 12822 | -0.064 | -0.3144 | No | ||
| 71 | NEUROG3 | 6980451 | 13322 | -0.083 | -0.3395 | No | ||
| 72 | PPOX | 2640678 3850102 | 13525 | -0.091 | -0.3483 | No | ||
| 73 | HNRPR | 2320440 2900601 4610008 | 13599 | -0.094 | -0.3501 | No | ||
| 74 | PLA2G2E | 4060128 | 13750 | -0.102 | -0.3559 | No | ||
| 75 | TIAL1 | 4150048 6510605 | 13794 | -0.104 | -0.3558 | No | ||
| 76 | POU2F1 | 70577 430373 4850324 5910056 | 14174 | -0.127 | -0.3734 | No | ||
| 77 | F2 | 5720280 | 14186 | -0.127 | -0.3711 | No | ||
| 78 | LBX1 | 1300164 4060204 | 14204 | -0.129 | -0.3690 | No | ||
| 79 | MDK | 3840020 | 14580 | -0.158 | -0.3857 | Yes | ||
| 80 | RFX5 | 5420128 | 14589 | -0.159 | -0.3824 | Yes | ||
| 81 | PPRC1 | 1740441 | 14616 | -0.161 | -0.3801 | Yes | ||
| 82 | PLCB3 | 4670402 | 14669 | -0.166 | -0.3791 | Yes | ||
| 83 | MDM4 | 4070504 4780008 | 14699 | -0.168 | -0.3768 | Yes | ||
| 84 | DUSP8 | 6650039 | 14730 | -0.171 | -0.3745 | Yes | ||
| 85 | PTPN5 | 2350161 | 14758 | -0.174 | -0.3720 | Yes | ||
| 86 | EPHA2 | 5890056 | 14788 | -0.177 | -0.3695 | Yes | ||
| 87 | BAI2 | 380632 630113 6650170 | 14795 | -0.178 | -0.3657 | Yes | ||
| 88 | GRIK3 | 6380592 | 14904 | -0.188 | -0.3672 | Yes | ||
| 89 | FHL3 | 1090369 2650017 | 15118 | -0.212 | -0.3738 | Yes | ||
| 90 | MLLT10 | 1410288 3290142 | 15191 | -0.222 | -0.3726 | Yes | ||
| 91 | P4HA1 | 2630692 | 15489 | -0.267 | -0.3826 | Yes | ||
| 92 | GALNT2 | 2260279 4060239 | 15495 | -0.268 | -0.3767 | Yes | ||
| 93 | TRIM33 | 580619 2230280 3990433 6200747 | 15643 | -0.293 | -0.3779 | Yes | ||
| 94 | PACSIN3 | 4540369 4540400 5360037 6940129 | 15647 | -0.293 | -0.3713 | Yes | ||
| 95 | HSD11B1 | 450066 5550408 | 15739 | -0.309 | -0.3691 | Yes | ||
| 96 | KRTCAP2 | 5220040 | 15806 | -0.322 | -0.3653 | Yes | ||
| 97 | LCK | 3360142 | 15886 | -0.337 | -0.3618 | Yes | ||
| 98 | TRIM46 | 3520609 4590010 | 15889 | -0.338 | -0.3541 | Yes | ||
| 99 | BRDT | 3940102 | 15913 | -0.342 | -0.3475 | Yes | ||
| 100 | EPHB2 | 1090288 1410377 2100541 | 16175 | -0.403 | -0.3523 | Yes | ||
| 101 | GTF2H1 | 2570746 2650148 | 16213 | -0.412 | -0.3449 | Yes | ||
| 102 | CASQ1 | 6290035 7000707 | 16228 | -0.416 | -0.3361 | Yes | ||
| 103 | PHC2 | 1430433 | 16256 | -0.424 | -0.3278 | Yes | ||
| 104 | NCDN | 50040 1980739 3520603 | 16265 | -0.428 | -0.3183 | Yes | ||
| 105 | DNAJB12 | 2030148 | 16307 | -0.439 | -0.3105 | Yes | ||
| 106 | YARS | 70739 | 16312 | -0.441 | -0.3005 | Yes | ||
| 107 | MOV10 | 50673 430577 3870280 | 16442 | -0.478 | -0.2965 | Yes | ||
| 108 | NR1H3 | 4810215 5570292 | 16500 | -0.491 | -0.2883 | Yes | ||
| 109 | ARID1A | 2630022 1690551 4810110 | 16639 | -0.539 | -0.2834 | Yes | ||
| 110 | SESN2 | 2370019 | 16670 | -0.550 | -0.2723 | Yes | ||
| 111 | PHF13 | 450435 | 16748 | -0.572 | -0.2633 | Yes | ||
| 112 | ATP1A1 | 5670451 | 16905 | -0.623 | -0.2574 | Yes | ||
| 113 | GFRA1 | 3610152 | 17009 | -0.668 | -0.2476 | Yes | ||
| 114 | FDPS | 60022 | 17543 | -0.917 | -0.2554 | Yes | ||
| 115 | ARHGEF2 | 3360577 | 17626 | -0.956 | -0.2378 | Yes | ||
| 116 | TNNI2 | 5910739 | 17636 | -0.963 | -0.2161 | Yes | ||
| 117 | TPM3 | 670600 3170296 5670167 6650471 | 17718 | -1.001 | -0.1975 | Yes | ||
| 118 | OTUB1 | 610551 | 17746 | -1.017 | -0.1755 | Yes | ||
| 119 | HMGN2 | 3140091 | 17854 | -1.096 | -0.1561 | Yes | ||
| 120 | UPF2 | 4810471 | 17961 | -1.173 | -0.1349 | Yes | ||
| 121 | ARFIP2 | 1850301 4670180 | 18032 | -1.232 | -0.1103 | Yes | ||
| 122 | LRRC20 | 6290373 | 18083 | -1.278 | -0.0836 | Yes | ||
| 123 | RHOG | 6760575 | 18173 | -1.375 | -0.0568 | Yes | ||
| 124 | FKBP2 | 2320025 | 18215 | -1.442 | -0.0258 | Yes | ||
| 125 | PTPRF | 1770528 3190044 | 18502 | -2.063 | 0.0062 | Yes |

