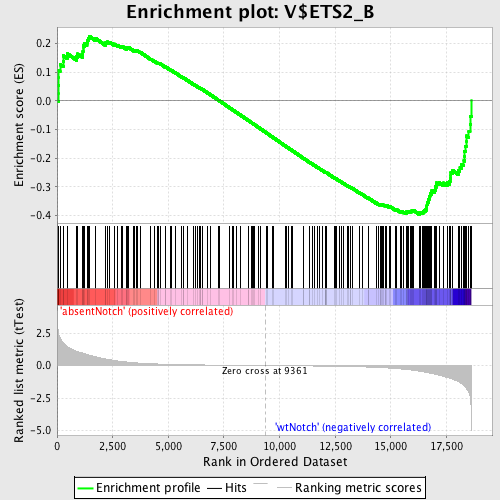
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$ETS2_B |
| Enrichment Score (ES) | -0.39672014 |
| Normalized Enrichment Score (NES) | -1.2403876 |
| Nominal p-value | 0.04368932 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TYROBP | 2230020 | 50 | 2.584 | 0.0261 | No | ||
| 2 | NR1D1 | 2360471 770746 6590204 | 54 | 2.528 | 0.0540 | No | ||
| 3 | S100A10 | 6350010 | 71 | 2.418 | 0.0801 | No | ||
| 4 | IMPDH1 | 3190735 7050546 | 77 | 2.354 | 0.1060 | No | ||
| 5 | USP3 | 2060332 | 133 | 2.154 | 0.1270 | No | ||
| 6 | VCL | 4120487 | 273 | 1.734 | 0.1388 | No | ||
| 7 | LST1 | 5420372 | 288 | 1.714 | 0.1571 | No | ||
| 8 | CKB | 4560632 | 454 | 1.473 | 0.1646 | No | ||
| 9 | SPATA6 | 4590373 | 878 | 1.097 | 0.1538 | No | ||
| 10 | CTCF | 5340017 | 920 | 1.070 | 0.1635 | No | ||
| 11 | ELMO1 | 110338 1190451 1580148 1850609 2350706 2370369 2450129 3840113 4150184 4280079 6290066 6380435 6770494 7100180 | 1133 | 0.970 | 0.1628 | No | ||
| 12 | SESTD1 | 1090333 3780593 | 1159 | 0.957 | 0.1721 | No | ||
| 13 | UTX | 2900017 6770452 | 1170 | 0.954 | 0.1822 | No | ||
| 14 | S100A9 | 7050528 | 1172 | 0.953 | 0.1927 | No | ||
| 15 | RGS12 | 1190082 5390369 | 1224 | 0.916 | 0.2002 | No | ||
| 16 | CALM2 | 6620463 | 1344 | 0.852 | 0.2032 | No | ||
| 17 | ARHGAP4 | 1940022 | 1375 | 0.836 | 0.2109 | No | ||
| 18 | VDAC2 | 7000215 | 1416 | 0.820 | 0.2178 | No | ||
| 19 | RAC1 | 4810687 | 1437 | 0.806 | 0.2257 | No | ||
| 20 | IGF1 | 1990193 3130377 3290280 | 1730 | 0.674 | 0.2174 | No | ||
| 21 | DLGAP4 | 4570672 5690161 | 2175 | 0.506 | 0.1989 | No | ||
| 22 | CITED2 | 5670114 5130088 | 2190 | 0.502 | 0.2037 | No | ||
| 23 | TCF12 | 3610324 7000156 | 2257 | 0.484 | 0.2055 | No | ||
| 24 | TCERG1 | 2350551 4540397 5220746 | 2366 | 0.445 | 0.2046 | No | ||
| 25 | ATP5H | 670438 | 2566 | 0.394 | 0.1982 | No | ||
| 26 | TRPC4AP | 840520 1660402 | 2708 | 0.359 | 0.1946 | No | ||
| 27 | GNG11 | 4010142 | 2887 | 0.319 | 0.1885 | No | ||
| 28 | FYN | 2100468 4760520 4850687 | 2920 | 0.311 | 0.1902 | No | ||
| 29 | PITPNB | 5130039 | 3117 | 0.272 | 0.1826 | No | ||
| 30 | BCL2L2 | 2760692 6770739 | 3126 | 0.271 | 0.1852 | No | ||
| 31 | RASA3 | 2060465 | 3165 | 0.262 | 0.1860 | No | ||
| 32 | CANX | 4760402 | 3220 | 0.252 | 0.1859 | No | ||
| 33 | GPR44 | 4050452 | 3451 | 0.219 | 0.1759 | No | ||
| 34 | DAB2IP | 3170064 6200441 | 3493 | 0.212 | 0.1760 | No | ||
| 35 | E2F3 | 50162 460180 | 3567 | 0.203 | 0.1743 | No | ||
| 36 | FURIN | 4120168 | 3599 | 0.199 | 0.1748 | No | ||
| 37 | PCDH7 | 450398 4150040 | 3734 | 0.185 | 0.1696 | No | ||
| 38 | LDB2 | 5670441 6110170 | 4215 | 0.140 | 0.1451 | No | ||
| 39 | WNT9A | 3830711 | 4366 | 0.129 | 0.1384 | No | ||
| 40 | CNKSR1 | 730528 3390672 | 4509 | 0.119 | 0.1321 | No | ||
| 41 | DPPA4 | 6860692 6900164 | 4537 | 0.118 | 0.1319 | No | ||
| 42 | KLF12 | 1660095 4810288 5340546 6520286 | 4562 | 0.117 | 0.1319 | No | ||
| 43 | CENTD2 | 60408 2510156 6100494 | 4628 | 0.112 | 0.1296 | No | ||
| 44 | PPP1R12A | 4070066 4590162 | 4851 | 0.100 | 0.1187 | No | ||
| 45 | LRMP | 6290193 | 4892 | 0.098 | 0.1176 | No | ||
| 46 | TUSC3 | 1690411 | 5114 | 0.089 | 0.1066 | No | ||
| 47 | FUT10 | 6510603 | 5155 | 0.087 | 0.1054 | No | ||
| 48 | MMP3 | 2650368 2970056 | 5321 | 0.080 | 0.0973 | No | ||
| 49 | PAK3 | 4210136 | 5602 | 0.071 | 0.0829 | No | ||
| 50 | DLC1 | 1090632 6450594 | 5658 | 0.069 | 0.0807 | No | ||
| 51 | SLC39A11 | 5860687 | 5702 | 0.067 | 0.0791 | No | ||
| 52 | STARD13 | 540722 | 5856 | 0.063 | 0.0715 | No | ||
| 53 | HOXA1 | 1190524 5420142 | 6140 | 0.055 | 0.0568 | No | ||
| 54 | SHC3 | 2690398 | 6209 | 0.053 | 0.0537 | No | ||
| 55 | CLSPN | 2190239 2630687 5550632 | 6311 | 0.051 | 0.0488 | No | ||
| 56 | SP3 | 3840338 | 6387 | 0.049 | 0.0453 | No | ||
| 57 | LOH11CR2A | 4480403 | 6424 | 0.048 | 0.0438 | No | ||
| 58 | CAMK2A | 1740333 1940112 4150292 | 6459 | 0.047 | 0.0425 | No | ||
| 59 | EVX1 | 1980471 | 6517 | 0.046 | 0.0399 | No | ||
| 60 | LOXL3 | 4010168 | 6519 | 0.046 | 0.0404 | No | ||
| 61 | ATOH8 | 4540204 | 6769 | 0.041 | 0.0273 | No | ||
| 62 | PGF | 4810593 | 6879 | 0.038 | 0.0218 | No | ||
| 63 | C1QTNF5 | 1690402 | 7246 | 0.031 | 0.0023 | No | ||
| 64 | APBA1 | 3140487 | 7278 | 0.031 | 0.0010 | No | ||
| 65 | NPAS2 | 770670 | 7747 | 0.023 | -0.0241 | No | ||
| 66 | TIMP4 | 6110433 | 7878 | 0.021 | -0.0310 | No | ||
| 67 | PAX6 | 1190025 | 7921 | 0.021 | -0.0330 | No | ||
| 68 | GPR81 | 3190541 | 8056 | 0.019 | -0.0401 | No | ||
| 69 | TREML1 | 2510528 | 8225 | 0.016 | -0.0490 | No | ||
| 70 | TWIST1 | 5130619 | 8583 | 0.011 | -0.0682 | No | ||
| 71 | AP2B1 | 1450184 2370148 3520139 4850593 | 8589 | 0.011 | -0.0684 | No | ||
| 72 | MYH10 | 2640673 | 8745 | 0.008 | -0.0767 | No | ||
| 73 | STRBP | 4210594 5360239 | 8793 | 0.008 | -0.0792 | No | ||
| 74 | ACTR3 | 1400497 | 8811 | 0.007 | -0.0800 | No | ||
| 75 | CPNE8 | 7040039 | 8881 | 0.007 | -0.0837 | No | ||
| 76 | CTHRC1 | 4150020 | 9070 | 0.004 | -0.0938 | No | ||
| 77 | LRP5 | 2100397 3170484 | 9158 | 0.003 | -0.0985 | No | ||
| 78 | RHOV | 5860100 | 9405 | -0.001 | -0.1119 | No | ||
| 79 | DOCK4 | 5910102 | 9443 | -0.001 | -0.1139 | No | ||
| 80 | VIP | 2850647 | 9666 | -0.004 | -0.1259 | No | ||
| 81 | ANGPT1 | 3990368 5130288 6770035 | 9731 | -0.005 | -0.1293 | No | ||
| 82 | ZNF205 | 6900022 | 10257 | -0.012 | -0.1576 | No | ||
| 83 | ZNF35 | 670156 | 10327 | -0.014 | -0.1612 | No | ||
| 84 | IL13 | 7040021 | 10422 | -0.015 | -0.1662 | No | ||
| 85 | CRK | 1230162 4780128 | 10536 | -0.017 | -0.1721 | No | ||
| 86 | ARHGEF12 | 3990195 | 10578 | -0.017 | -0.1741 | No | ||
| 87 | TYRO3 | 3130193 | 11091 | -0.026 | -0.2016 | No | ||
| 88 | DIXDC1 | 6980435 | 11092 | -0.026 | -0.2013 | No | ||
| 89 | PTGIR | 2100563 | 11340 | -0.030 | -0.2144 | No | ||
| 90 | SPARC | 1690086 | 11350 | -0.030 | -0.2146 | No | ||
| 91 | FLI1 | 3990142 | 11473 | -0.032 | -0.2208 | No | ||
| 92 | DOK1 | 1050279 2680112 5220273 | 11547 | -0.034 | -0.2244 | No | ||
| 93 | TEAD3 | 2320142 | 11693 | -0.037 | -0.2319 | No | ||
| 94 | ITPR2 | 5360128 | 11794 | -0.039 | -0.2368 | No | ||
| 95 | REST | 1500471 | 11927 | -0.041 | -0.2436 | No | ||
| 96 | IL12B | 460008 | 12044 | -0.044 | -0.2494 | No | ||
| 97 | TREX2 | 4920707 | 12048 | -0.044 | -0.2490 | No | ||
| 98 | RTKN | 5910253 6380082 | 12064 | -0.044 | -0.2494 | No | ||
| 99 | ANGPTL1 | 4570528 5290301 6400541 | 12092 | -0.045 | -0.2503 | No | ||
| 100 | HOXA10 | 6110397 7100458 | 12108 | -0.045 | -0.2506 | No | ||
| 101 | FLNC | 2360048 5050270 | 12481 | -0.055 | -0.2702 | No | ||
| 102 | RNF12 | 6860717 | 12512 | -0.055 | -0.2712 | No | ||
| 103 | LYN | 6040600 | 12550 | -0.056 | -0.2726 | No | ||
| 104 | VASP | 7050500 | 12671 | -0.059 | -0.2785 | No | ||
| 105 | APOB48R | 2810070 | 12775 | -0.063 | -0.2833 | No | ||
| 106 | CAST | 3850082 5420671 6200156 | 12867 | -0.065 | -0.2876 | No | ||
| 107 | HOXC4 | 1940193 | 13066 | -0.073 | -0.2975 | No | ||
| 108 | EFNB3 | 5570594 | 13115 | -0.075 | -0.2993 | No | ||
| 109 | RNF128 | 1230300 5670273 | 13167 | -0.077 | -0.3012 | No | ||
| 110 | OLFM1 | 3360161 5570066 | 13201 | -0.078 | -0.3021 | No | ||
| 111 | ACTR2 | 1170332 | 13294 | -0.082 | -0.3062 | No | ||
| 112 | LIFR | 3360068 3830102 | 13607 | -0.095 | -0.3221 | No | ||
| 113 | HAPLN1 | 580398 | 13724 | -0.100 | -0.3272 | No | ||
| 114 | MAP3K11 | 7000039 | 13987 | -0.115 | -0.3402 | No | ||
| 115 | GMPR2 | 4920204 | 13989 | -0.115 | -0.3389 | No | ||
| 116 | FGFR4 | 2650072 | 14336 | -0.138 | -0.3562 | No | ||
| 117 | NT5C3 | 1570075 | 14462 | -0.147 | -0.3613 | No | ||
| 118 | RAB20 | 2640601 | 14513 | -0.152 | -0.3623 | No | ||
| 119 | MYCT1 | 5670044 | 14517 | -0.152 | -0.3608 | No | ||
| 120 | LGALS4 | 2260215 3390338 | 14561 | -0.157 | -0.3614 | No | ||
| 121 | PPRC1 | 1740441 | 14616 | -0.161 | -0.3625 | No | ||
| 122 | SEMA4C | 1980315 | 14668 | -0.166 | -0.3635 | No | ||
| 123 | PSIP1 | 1780082 3190435 5050594 | 14740 | -0.172 | -0.3654 | No | ||
| 124 | SPI1 | 1410397 | 14750 | -0.173 | -0.3639 | No | ||
| 125 | EPHA2 | 5890056 | 14788 | -0.177 | -0.3640 | No | ||
| 126 | CACNA1D | 1740315 4730731 | 14960 | -0.194 | -0.3711 | No | ||
| 127 | TAF5 | 3450288 5890193 6860435 | 14976 | -0.195 | -0.3698 | No | ||
| 128 | CDH5 | 5340487 | 15220 | -0.225 | -0.3804 | No | ||
| 129 | DGKZ | 3460041 6400129 | 15274 | -0.233 | -0.3807 | No | ||
| 130 | DPP3 | 2680725 3780670 | 15432 | -0.255 | -0.3864 | No | ||
| 131 | C1QTNF6 | 2030592 | 15490 | -0.267 | -0.3865 | No | ||
| 132 | MAP4K1 | 4060692 | 15584 | -0.284 | -0.3884 | No | ||
| 133 | ARHGDIB | 4730433 | 15694 | -0.301 | -0.3910 | No | ||
| 134 | IRAK4 | 5390156 6130594 | 15696 | -0.302 | -0.3876 | No | ||
| 135 | ORC4L | 4230538 5550288 | 15761 | -0.313 | -0.3876 | No | ||
| 136 | EIF5A | 1500059 5290022 6370026 | 15794 | -0.319 | -0.3858 | No | ||
| 137 | CTSW | 360368 | 15883 | -0.337 | -0.3868 | No | ||
| 138 | SSBP4 | 2570735 | 15909 | -0.342 | -0.3844 | No | ||
| 139 | TMOD3 | 840088 | 15977 | -0.357 | -0.3841 | No | ||
| 140 | CORO1C | 4570397 | 16012 | -0.364 | -0.3819 | No | ||
| 141 | ESM1 | 1410594 | 16287 | -0.434 | -0.3919 | Yes | ||
| 142 | SLC31A1 | 4590161 6940154 | 16322 | -0.444 | -0.3888 | Yes | ||
| 143 | LYL1 | 2900242 | 16430 | -0.476 | -0.3893 | Yes | ||
| 144 | CA4 | 130100 | 16475 | -0.486 | -0.3863 | Yes | ||
| 145 | BZW2 | 940079 | 16510 | -0.495 | -0.3826 | Yes | ||
| 146 | TLN1 | 6590411 | 16573 | -0.517 | -0.3802 | Yes | ||
| 147 | ATP8B2 | 6590403 | 16597 | -0.525 | -0.3756 | Yes | ||
| 148 | ITGB7 | 4670619 | 16610 | -0.529 | -0.3704 | Yes | ||
| 149 | IKBKB | 6840072 | 16623 | -0.532 | -0.3651 | Yes | ||
| 150 | CDKN1B | 3800025 6450044 | 16626 | -0.533 | -0.3593 | Yes | ||
| 151 | LAT | 3170025 | 16657 | -0.545 | -0.3548 | Yes | ||
| 152 | EGR1 | 4610347 | 16691 | -0.558 | -0.3504 | Yes | ||
| 153 | SIPA1 | 5220687 | 16698 | -0.559 | -0.3445 | Yes | ||
| 154 | CUEDC1 | 1660463 | 16723 | -0.566 | -0.3395 | Yes | ||
| 155 | SLC9A3R1 | 6020373 | 16735 | -0.568 | -0.3338 | Yes | ||
| 156 | GALE | 3610400 6290072 | 16761 | -0.577 | -0.3287 | Yes | ||
| 157 | IL7R | 2060088 6380500 | 16767 | -0.579 | -0.3226 | Yes | ||
| 158 | RGS3 | 60670 540736 1340180 1500369 3390735 4010131 4610402 6380114 | 16834 | -0.602 | -0.3194 | Yes | ||
| 159 | GTF2B | 6860685 | 16846 | -0.604 | -0.3133 | Yes | ||
| 160 | TGIF2 | 3840364 4050520 | 16978 | -0.655 | -0.3131 | Yes | ||
| 161 | MCTS1 | 6130133 | 17002 | -0.666 | -0.3070 | Yes | ||
| 162 | CAMK1D | 1660100 2350309 | 17007 | -0.667 | -0.2998 | Yes | ||
| 163 | ZHX2 | 2900452 | 17036 | -0.680 | -0.2937 | Yes | ||
| 164 | RAB11A | 5360079 | 17046 | -0.686 | -0.2866 | Yes | ||
| 165 | NEDD8 | 2350722 | 17168 | -0.733 | -0.2850 | Yes | ||
| 166 | CLTA | 450010 | 17369 | -0.819 | -0.2867 | Yes | ||
| 167 | L3MBTL2 | 6020341 | 17534 | -0.910 | -0.2855 | Yes | ||
| 168 | TNNI2 | 5910739 | 17636 | -0.963 | -0.2802 | Yes | ||
| 169 | ARHGEF6 | 4780164 6380450 | 17666 | -0.984 | -0.2708 | Yes | ||
| 170 | PSEN1 | 130403 2030647 6100603 | 17694 | -0.996 | -0.2612 | Yes | ||
| 171 | DMTF1 | 5910047 1570497 2480100 6620239 | 17700 | -0.998 | -0.2504 | Yes | ||
| 172 | BAZ2A | 730184 | 17764 | -1.024 | -0.2424 | Yes | ||
| 173 | PPP1R9B | 3130619 | 18031 | -1.227 | -0.2432 | Yes | ||
| 174 | RNF40 | 3610397 | 18108 | -1.306 | -0.2328 | Yes | ||
| 175 | CREB3 | 5220408 | 18188 | -1.396 | -0.2215 | Yes | ||
| 176 | AAMP | 2640059 | 18287 | -1.551 | -0.2096 | Yes | ||
| 177 | LCP1 | 4280397 | 18306 | -1.578 | -0.1930 | Yes | ||
| 178 | BCL3 | 3990440 | 18315 | -1.592 | -0.1757 | Yes | ||
| 179 | FGR | 3780047 | 18351 | -1.662 | -0.1591 | Yes | ||
| 180 | GCNT2 | 6400162 2370609 2760309 4540465 6100280 | 18388 | -1.733 | -0.1417 | Yes | ||
| 181 | MAPKAPK2 | 2850500 | 18395 | -1.746 | -0.1226 | Yes | ||
| 182 | GIT2 | 6040039 6770181 | 18509 | -2.094 | -0.1054 | Yes | ||
| 183 | RASAL1 | 3710112 | 18559 | -2.379 | -0.0816 | Yes | ||
| 184 | GNB1L | 3840368 5270215 | 18583 | -2.662 | -0.0532 | Yes | ||
| 185 | POU2AF1 | 5690242 | 18615 | -4.937 | 0.0001 | Yes |

