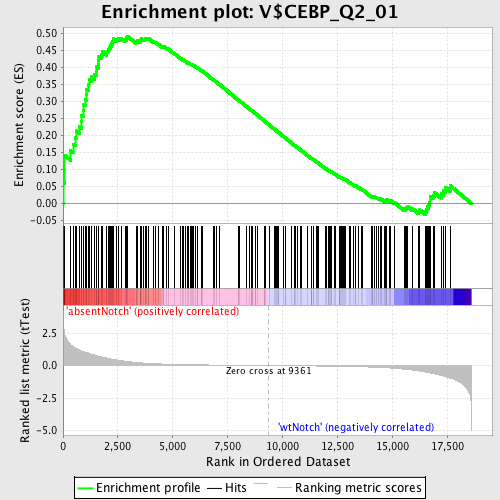
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$CEBP_Q2_01 |
| Enrichment Score (ES) | 0.4928466 |
| Normalized Enrichment Score (NES) | 1.4945948 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17188849 |
| FWER p-Value | 0.922 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC40A1 | 2370286 | 5 | 3.698 | 0.0639 | Yes | ||
| 2 | PER1 | 3290315 | 76 | 2.373 | 0.1013 | Yes | ||
| 3 | H3F3B | 1410300 | 78 | 2.349 | 0.1420 | Yes | ||
| 4 | FOXP1 | 6510433 | 341 | 1.604 | 0.1556 | Yes | ||
| 5 | TBXAS1 | 5050053 6900286 | 457 | 1.466 | 0.1748 | Yes | ||
| 6 | STMN1 | 1990717 | 548 | 1.364 | 0.1936 | Yes | ||
| 7 | UBE2E2 | 610053 | 596 | 1.314 | 0.2138 | Yes | ||
| 8 | BAT2 | 6650435 | 750 | 1.196 | 0.2263 | Yes | ||
| 9 | NFIX | 2450152 | 835 | 1.136 | 0.2414 | Yes | ||
| 10 | ARF6 | 3520026 | 843 | 1.123 | 0.2605 | Yes | ||
| 11 | TOB1 | 4150138 | 937 | 1.057 | 0.2738 | Yes | ||
| 12 | ETV5 | 110017 | 949 | 1.049 | 0.2914 | Yes | ||
| 13 | SLIT3 | 7100132 | 1012 | 1.015 | 0.3057 | Yes | ||
| 14 | CDKN1C | 6520577 | 1066 | 0.999 | 0.3201 | Yes | ||
| 15 | EIF4A2 | 1170494 1740711 2850504 | 1076 | 0.997 | 0.3369 | Yes | ||
| 16 | RHOB | 1500309 | 1148 | 0.964 | 0.3498 | Yes | ||
| 17 | DYRK3 | 1090014 | 1180 | 0.947 | 0.3645 | Yes | ||
| 18 | CCL8 | 3870010 | 1283 | 0.881 | 0.3743 | Yes | ||
| 19 | CCL3 | 2810092 | 1448 | 0.800 | 0.3793 | Yes | ||
| 20 | MAP2K6 | 1230056 2940204 | 1511 | 0.770 | 0.3893 | Yes | ||
| 21 | RUVBL2 | 1190377 | 1519 | 0.768 | 0.4022 | Yes | ||
| 22 | WNT6 | 4570364 | 1611 | 0.723 | 0.4098 | Yes | ||
| 23 | PRDX3 | 1690035 | 1625 | 0.717 | 0.4215 | Yes | ||
| 24 | SNX12 | 4060026 | 1633 | 0.711 | 0.4335 | Yes | ||
| 25 | ETV6 | 610524 | 1750 | 0.666 | 0.4388 | Yes | ||
| 26 | BCL6 | 940100 | 1780 | 0.649 | 0.4485 | Yes | ||
| 27 | MPP6 | 6020148 | 1998 | 0.568 | 0.4465 | Yes | ||
| 28 | NFE2L2 | 2810619 3390162 | 2047 | 0.552 | 0.4535 | Yes | ||
| 29 | NEO1 | 1990154 | 2096 | 0.533 | 0.4602 | Yes | ||
| 30 | CFL2 | 380239 6200368 | 2164 | 0.509 | 0.4653 | Yes | ||
| 31 | CLASP1 | 6860279 | 2207 | 0.498 | 0.4717 | Yes | ||
| 32 | TCF12 | 3610324 7000156 | 2257 | 0.484 | 0.4775 | Yes | ||
| 33 | CYR61 | 1240408 5290026 4120452 6550008 | 2276 | 0.480 | 0.4848 | Yes | ||
| 34 | THRA | 6770341 110164 1990600 | 2433 | 0.427 | 0.4837 | Yes | ||
| 35 | LUZP1 | 940075 4280458 6180070 | 2526 | 0.403 | 0.4858 | Yes | ||
| 36 | ABHD6 | 630717 | 2641 | 0.375 | 0.4861 | Yes | ||
| 37 | ATAD2 | 870242 2260687 | 2821 | 0.334 | 0.4822 | Yes | ||
| 38 | MKNK2 | 1240020 5270092 | 2873 | 0.321 | 0.4850 | Yes | ||
| 39 | FOXP3 | 940707 1850692 | 2885 | 0.319 | 0.4899 | Yes | ||
| 40 | EPHB6 | 5860128 | 2930 | 0.308 | 0.4928 | Yes | ||
| 41 | ZFP36L1 | 2510138 4120048 | 3322 | 0.236 | 0.4757 | No | ||
| 42 | SPRED1 | 6940706 | 3357 | 0.231 | 0.4779 | No | ||
| 43 | STC1 | 360161 | 3377 | 0.228 | 0.4808 | No | ||
| 44 | FBXO40 | 2190286 5670253 | 3509 | 0.210 | 0.4774 | No | ||
| 45 | MAP3K3 | 610685 | 3541 | 0.206 | 0.4793 | No | ||
| 46 | TBR1 | 5130053 | 3548 | 0.205 | 0.4825 | No | ||
| 47 | HIST1H1C | 3870603 | 3556 | 0.204 | 0.4857 | No | ||
| 48 | FGA | 4210056 5220239 | 3653 | 0.193 | 0.4838 | No | ||
| 49 | PCDH7 | 450398 4150040 | 3734 | 0.185 | 0.4827 | No | ||
| 50 | SYNCRIP | 1690195 3140113 4670279 | 3755 | 0.182 | 0.4848 | No | ||
| 51 | FMO2 | 3850338 | 3783 | 0.177 | 0.4864 | No | ||
| 52 | ADAMTS2 | 2650025 6370133 6840670 | 3869 | 0.169 | 0.4847 | No | ||
| 53 | JUN | 840170 | 3909 | 0.166 | 0.4854 | No | ||
| 54 | ASCL2 | 1400273 | 4104 | 0.148 | 0.4775 | No | ||
| 55 | SPTLC2 | 6840324 520017 | 4196 | 0.141 | 0.4750 | No | ||
| 56 | CRIM1 | 130491 | 4343 | 0.131 | 0.4693 | No | ||
| 57 | IL1R2 | 1410600 | 4528 | 0.118 | 0.4614 | No | ||
| 58 | KLF12 | 1660095 4810288 5340546 6520286 | 4562 | 0.117 | 0.4616 | No | ||
| 59 | KLF5 | 3840348 | 4594 | 0.115 | 0.4620 | No | ||
| 60 | WEE1 | 3390070 | 4707 | 0.107 | 0.4577 | No | ||
| 61 | PCTP | 6980324 | 4782 | 0.103 | 0.4555 | No | ||
| 62 | SOX5 | 2370576 2900167 3190128 5050528 | 5064 | 0.091 | 0.4418 | No | ||
| 63 | BNC2 | 4810603 | 5345 | 0.079 | 0.4280 | No | ||
| 64 | MBNL1 | 2640762 7100048 | 5455 | 0.075 | 0.4234 | No | ||
| 65 | ASGR2 | 3360463 | 5467 | 0.075 | 0.4241 | No | ||
| 66 | EDG8 | 1990441 | 5595 | 0.071 | 0.4185 | No | ||
| 67 | FIGN | 7000601 | 5689 | 0.068 | 0.4146 | No | ||
| 68 | SPRY4 | 1570594 | 5721 | 0.067 | 0.4141 | No | ||
| 69 | CITED1 | 2350670 | 5819 | 0.064 | 0.4099 | No | ||
| 70 | LMO4 | 3800746 | 5835 | 0.064 | 0.4102 | No | ||
| 71 | DMD | 1740041 3990332 | 5875 | 0.062 | 0.4092 | No | ||
| 72 | ESR1 | 4060372 5860193 | 5942 | 0.060 | 0.4066 | No | ||
| 73 | ASAH2 | 3130452 | 6036 | 0.057 | 0.4026 | No | ||
| 74 | TNFSF15 | 360341 | 6110 | 0.055 | 0.3996 | No | ||
| 75 | HOXA1 | 1190524 5420142 | 6140 | 0.055 | 0.3990 | No | ||
| 76 | SLC6A15 | 5290176 6040017 | 6293 | 0.051 | 0.3916 | No | ||
| 77 | SEMA6D | 4050324 5860138 6350307 | 6321 | 0.051 | 0.3910 | No | ||
| 78 | TRIB1 | 2320435 | 6344 | 0.050 | 0.3907 | No | ||
| 79 | PPL | 2360072 | 6839 | 0.039 | 0.3646 | No | ||
| 80 | CHD2 | 4070039 | 6889 | 0.038 | 0.3626 | No | ||
| 81 | PRKG1 | 4120671 | 7010 | 0.036 | 0.3567 | No | ||
| 82 | CGN | 5890139 6370167 6400537 | 7112 | 0.034 | 0.3518 | No | ||
| 83 | IL27 | 1990324 | 7973 | 0.020 | 0.3055 | No | ||
| 84 | RBM4 | 2360113 5080750 | 7989 | 0.020 | 0.3050 | No | ||
| 85 | RGR | 730156 6400368 | 8055 | 0.019 | 0.3018 | No | ||
| 86 | NRXN3 | 1400546 3360471 | 8337 | 0.014 | 0.2868 | No | ||
| 87 | PCDHB4 | 670184 | 8483 | 0.012 | 0.2792 | No | ||
| 88 | USP9X | 130435 2370735 3120338 | 8576 | 0.011 | 0.2744 | No | ||
| 89 | EFEMP1 | 5860324 | 8582 | 0.011 | 0.2743 | No | ||
| 90 | PHF16 | 50373 7200161 | 8617 | 0.010 | 0.2726 | No | ||
| 91 | F9 | 3190154 | 8633 | 0.010 | 0.2720 | No | ||
| 92 | MYH10 | 2640673 | 8745 | 0.008 | 0.2661 | No | ||
| 93 | SDPR | 3360292 | 8749 | 0.008 | 0.2661 | No | ||
| 94 | HOXA2 | 2120121 | 8839 | 0.007 | 0.2614 | No | ||
| 95 | AP1S2 | 780279 4280707 | 9161 | 0.003 | 0.2440 | No | ||
| 96 | CALD1 | 1770129 1940397 | 9164 | 0.003 | 0.2439 | No | ||
| 97 | WNT3 | 2360685 | 9218 | 0.002 | 0.2411 | No | ||
| 98 | MITF | 380056 | 9394 | -0.000 | 0.2316 | No | ||
| 99 | CRH | 3710301 | 9633 | -0.004 | 0.2188 | No | ||
| 100 | ADAMTS5 | 5890592 | 9682 | -0.004 | 0.2162 | No | ||
| 101 | ANGPT1 | 3990368 5130288 6770035 | 9731 | -0.005 | 0.2137 | No | ||
| 102 | SPRR1B | 1190458 | 9750 | -0.005 | 0.2128 | No | ||
| 103 | TENC1 | 1980403 7050575 | 9817 | -0.006 | 0.2093 | No | ||
| 104 | EYA1 | 1450278 5220390 | 9836 | -0.006 | 0.2085 | No | ||
| 105 | LRRTM3 | 4210021 | 10023 | -0.009 | 0.1985 | No | ||
| 106 | EBF2 | 940324 | 10125 | -0.011 | 0.1932 | No | ||
| 107 | ELAVL2 | 360181 | 10145 | -0.011 | 0.1924 | No | ||
| 108 | TLR8 | 1240092 | 10423 | -0.015 | 0.1776 | No | ||
| 109 | ADAM9 | 3360411 | 10547 | -0.017 | 0.1713 | No | ||
| 110 | CALR | 510446 | 10559 | -0.017 | 0.1710 | No | ||
| 111 | KIAA1715 | 1190551 | 10602 | -0.018 | 0.1690 | No | ||
| 112 | CLDN8 | 1770113 2320500 | 10680 | -0.019 | 0.1651 | No | ||
| 113 | TNFSF14 | 1450369 5550594 | 10830 | -0.021 | 0.1574 | No | ||
| 114 | PLA2G4A | 6380364 | 10846 | -0.021 | 0.1570 | No | ||
| 115 | HOXA5 | 6840026 | 11134 | -0.026 | 0.1419 | No | ||
| 116 | EGFR | 4920138 6480521 | 11300 | -0.029 | 0.1334 | No | ||
| 117 | GABRR2 | 4560739 | 11303 | -0.029 | 0.1338 | No | ||
| 118 | SERPINA7 | 2510537 | 11329 | -0.029 | 0.1330 | No | ||
| 119 | IL1F10 | 2260647 | 11390 | -0.031 | 0.1302 | No | ||
| 120 | MOSPD2 | 4010064 | 11408 | -0.031 | 0.1299 | No | ||
| 121 | ALDH1A2 | 2320301 | 11552 | -0.034 | 0.1227 | No | ||
| 122 | SLC12A1 | 3450017 6620133 6770148 | 11584 | -0.035 | 0.1216 | No | ||
| 123 | MPP5 | 2100148 5220632 | 11629 | -0.035 | 0.1198 | No | ||
| 124 | PAX8 | 5050301 5290164 | 11957 | -0.042 | 0.1028 | No | ||
| 125 | CBX4 | 70332 | 11982 | -0.043 | 0.1023 | No | ||
| 126 | SULF1 | 430575 | 12090 | -0.045 | 0.0972 | No | ||
| 127 | HOXA10 | 6110397 7100458 | 12108 | -0.045 | 0.0971 | No | ||
| 128 | LHX6 | 2360731 3360397 | 12159 | -0.046 | 0.0952 | No | ||
| 129 | DBH | 3610458 | 12168 | -0.047 | 0.0956 | No | ||
| 130 | PLCB2 | 360132 | 12232 | -0.048 | 0.0930 | No | ||
| 131 | RARB | 430139 1410138 | 12233 | -0.048 | 0.0938 | No | ||
| 132 | TNNC2 | 3840079 | 12351 | -0.051 | 0.0883 | No | ||
| 133 | FBXL14 | 4670315 | 12425 | -0.053 | 0.0853 | No | ||
| 134 | GPC4 | 5910408 | 12593 | -0.057 | 0.0772 | No | ||
| 135 | BMX | 50136 2940242 4540059 | 12597 | -0.057 | 0.0781 | No | ||
| 136 | PITX2 | 870537 2690139 | 12609 | -0.058 | 0.0785 | No | ||
| 137 | MYH4 | 1940102 | 12661 | -0.059 | 0.0767 | No | ||
| 138 | IL19 | 6040142 | 12672 | -0.059 | 0.0772 | No | ||
| 139 | CASK | 2340215 3290576 6770215 | 12729 | -0.061 | 0.0752 | No | ||
| 140 | LIPG | 1770040 | 12799 | -0.063 | 0.0726 | No | ||
| 141 | BDNF | 2940128 3520368 | 12822 | -0.064 | 0.0725 | No | ||
| 142 | PLCB1 | 2570050 7100102 | 12852 | -0.065 | 0.0720 | No | ||
| 143 | HOXC4 | 1940193 | 13066 | -0.073 | 0.0618 | No | ||
| 144 | ASGR1 | 1850088 | 13107 | -0.074 | 0.0609 | No | ||
| 145 | FGF14 | 2630390 3520075 6770048 | 13250 | -0.079 | 0.0545 | No | ||
| 146 | PPP1CB | 3130411 3780270 4150180 | 13311 | -0.082 | 0.0527 | No | ||
| 147 | GYS1 | 540154 | 13342 | -0.084 | 0.0525 | No | ||
| 148 | NRP2 | 4070400 5860041 6650446 | 13477 | -0.089 | 0.0468 | No | ||
| 149 | LEMD2 | 1940156 | 13577 | -0.093 | 0.0431 | No | ||
| 150 | DOCK3 | 3290273 3360497 2850068 | 13641 | -0.096 | 0.0413 | No | ||
| 151 | NFIL3 | 4070377 | 14075 | -0.120 | 0.0199 | No | ||
| 152 | ADH7 | 60465 | 14113 | -0.123 | 0.0200 | No | ||
| 153 | HOXB7 | 4540706 4610358 | 14123 | -0.123 | 0.0217 | No | ||
| 154 | PDE4D | 2470528 6660014 | 14192 | -0.128 | 0.0202 | No | ||
| 155 | PTEN | 3390064 | 14263 | -0.132 | 0.0187 | No | ||
| 156 | TNFSF13B | 1940438 6510441 | 14363 | -0.140 | 0.0157 | No | ||
| 157 | TESK2 | 6520524 | 14461 | -0.147 | 0.0130 | No | ||
| 158 | ARRDC3 | 4610390 | 14499 | -0.150 | 0.0136 | No | ||
| 159 | ERG | 50154 1770739 | 14665 | -0.166 | 0.0076 | No | ||
| 160 | ERF | 110551 1770278 | 14694 | -0.168 | 0.0090 | No | ||
| 161 | SPAG9 | 580497 2510180 4210685 | 14719 | -0.170 | 0.0106 | No | ||
| 162 | XKRX | 3800192 | 14741 | -0.172 | 0.0125 | No | ||
| 163 | XDH | 6840446 | 14884 | -0.186 | 0.0080 | No | ||
| 164 | FBXW7 | 4210338 7050280 | 14925 | -0.190 | 0.0091 | No | ||
| 165 | WNT5A | 840685 3120152 | 15104 | -0.211 | 0.0031 | No | ||
| 166 | APC | 3850484 5860722 | 15544 | -0.277 | -0.0159 | No | ||
| 167 | HOXD9 | 2570181 | 15618 | -0.288 | -0.0149 | No | ||
| 168 | HOXC6 | 3850133 4920736 | 15642 | -0.292 | -0.0111 | No | ||
| 169 | STC2 | 4920601 | 15717 | -0.306 | -0.0098 | No | ||
| 170 | TBX6 | 4730139 6840647 | 15933 | -0.346 | -0.0154 | No | ||
| 171 | CEPT1 | 1940047 | 16197 | -0.409 | -0.0226 | No | ||
| 172 | WNT10B | 510050 | 16257 | -0.424 | -0.0184 | No | ||
| 173 | DLX1 | 360168 | 16507 | -0.493 | -0.0234 | No | ||
| 174 | NEUROG1 | 380341 | 16574 | -0.517 | -0.0180 | No | ||
| 175 | DCN | 510332 5340026 5900711 6550092 | 16587 | -0.523 | -0.0096 | No | ||
| 176 | TSGA14 | 6660465 | 16666 | -0.547 | -0.0043 | No | ||
| 177 | TEGT | 1770538 | 16684 | -0.555 | 0.0044 | No | ||
| 178 | HAGH | 540358 | 16721 | -0.565 | 0.0122 | No | ||
| 179 | CUEDC1 | 1660463 | 16723 | -0.566 | 0.0220 | No | ||
| 180 | ADRB2 | 3290373 | 16860 | -0.609 | 0.0252 | No | ||
| 181 | MAP2K3 | 5570193 | 16919 | -0.629 | 0.0330 | No | ||
| 182 | RIN3 | 1940044 | 17226 | -0.758 | 0.0295 | No | ||
| 183 | YWHAG | 3780341 | 17341 | -0.804 | 0.0373 | No | ||
| 184 | HIVEP3 | 3990551 | 17444 | -0.861 | 0.0467 | No | ||
| 185 | CLN5 | 630767 | 17642 | -0.971 | 0.0528 | No |

