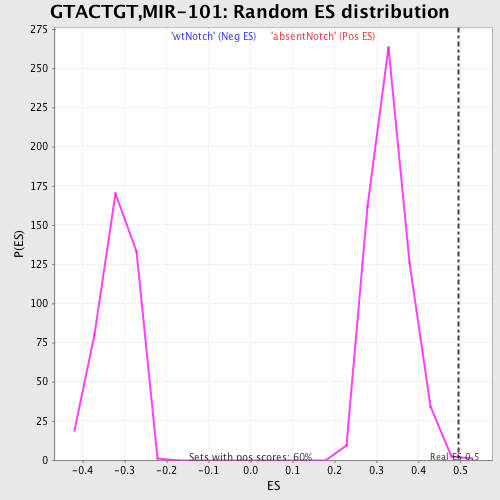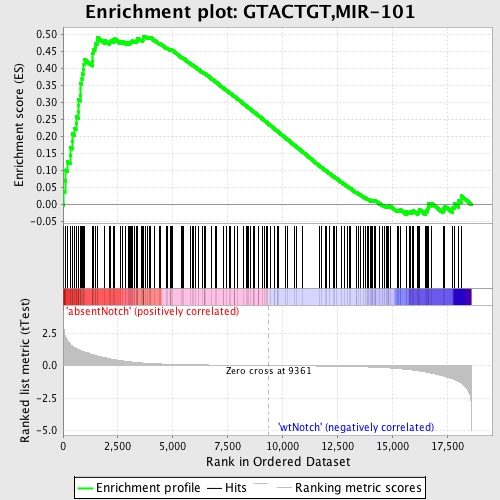
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | GTACTGT,MIR-101 |
| Enrichment Score (ES) | 0.49557617 |
| Normalized Enrichment Score (NES) | 1.4942629 |
| Nominal p-value | 0.0016750419 |
| FDR q-value | 0.16394278 |
| FWER p-Value | 0.922 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BCL2L11 | 780044 4200601 | 37 | 2.778 | 0.0395 | Yes | ||
| 2 | EMP1 | 4120438 | 87 | 2.294 | 0.0712 | Yes | ||
| 3 | DUSP1 | 6860121 | 125 | 2.179 | 0.1018 | Yes | ||
| 4 | WSB1 | 870563 | 209 | 1.911 | 0.1259 | Yes | ||
| 5 | RAP1B | 3710138 | 319 | 1.657 | 0.1447 | Yes | ||
| 6 | DOT1L | 3850068 | 342 | 1.603 | 0.1675 | Yes | ||
| 7 | AP3S1 | 5570044 | 417 | 1.515 | 0.1861 | Yes | ||
| 8 | GNB1 | 2120397 | 426 | 1.508 | 0.2083 | Yes | ||
| 9 | ZFAND3 | 2760504 | 534 | 1.375 | 0.2230 | Yes | ||
| 10 | STK4 | 2640152 | 591 | 1.319 | 0.2397 | Yes | ||
| 11 | CBL | 6380068 | 609 | 1.306 | 0.2583 | Yes | ||
| 12 | NACA | 520326 5890673 | 691 | 1.235 | 0.2724 | Yes | ||
| 13 | MTMR2 | 2260148 3990139 | 695 | 1.233 | 0.2907 | Yes | ||
| 14 | JDP2 | 2360500 | 701 | 1.230 | 0.3088 | Yes | ||
| 15 | APP | 2510053 | 778 | 1.180 | 0.3224 | Yes | ||
| 16 | GJA1 | 5220731 | 803 | 1.160 | 0.3384 | Yes | ||
| 17 | SLC19A2 | 6200750 | 804 | 1.160 | 0.3558 | Yes | ||
| 18 | DDX3X | 2190020 | 857 | 1.115 | 0.3696 | Yes | ||
| 19 | SPOP | 450035 | 869 | 1.104 | 0.3855 | Yes | ||
| 20 | CTCF | 5340017 | 920 | 1.070 | 0.3988 | Yes | ||
| 21 | ETV5 | 110017 | 949 | 1.049 | 0.4130 | Yes | ||
| 22 | SOCS2 | 4760692 | 988 | 1.024 | 0.4262 | Yes | ||
| 23 | PHLDA1 | 2450020 | 1332 | 0.856 | 0.4204 | Yes | ||
| 24 | SFRS5 | 3450176 6350008 | 1348 | 0.849 | 0.4323 | Yes | ||
| 25 | SLC30A7 | 2230463 | 1349 | 0.848 | 0.4450 | Yes | ||
| 26 | KPNB1 | 1690138 | 1386 | 0.833 | 0.4555 | Yes | ||
| 27 | SGPL1 | 4480059 | 1454 | 0.796 | 0.4638 | Yes | ||
| 28 | PURB | 5360138 | 1492 | 0.778 | 0.4734 | Yes | ||
| 29 | ICK | 1580746 3140021 | 1551 | 0.752 | 0.4815 | Yes | ||
| 30 | RANBP9 | 4670685 | 1577 | 0.741 | 0.4913 | Yes | ||
| 31 | CDK6 | 4920253 | 1888 | 0.610 | 0.4836 | Yes | ||
| 32 | PIP5K1C | 5890164 6510148 | 2130 | 0.521 | 0.4783 | Yes | ||
| 33 | FBN2 | 50435 | 2181 | 0.505 | 0.4831 | Yes | ||
| 34 | ZNF24 | 3830750 | 2284 | 0.477 | 0.4847 | Yes | ||
| 35 | UBE2D2 | 4590671 6510446 | 2361 | 0.448 | 0.4873 | Yes | ||
| 36 | RRM1 | 4150433 | 2613 | 0.383 | 0.4794 | Yes | ||
| 37 | CPEB2 | 4760338 | 2698 | 0.360 | 0.4803 | Yes | ||
| 38 | RXRB | 1780040 5340438 | 2850 | 0.327 | 0.4770 | Yes | ||
| 39 | PLXNB1 | 6220273 | 2969 | 0.299 | 0.4750 | Yes | ||
| 40 | RAP2C | 1690132 | 3024 | 0.288 | 0.4764 | Yes | ||
| 41 | ATXN1 | 5550156 | 3076 | 0.278 | 0.4778 | Yes | ||
| 42 | CEBPA | 5690519 | 3108 | 0.275 | 0.4802 | Yes | ||
| 43 | EZH2 | 6130605 6380524 | 3159 | 0.264 | 0.4815 | Yes | ||
| 44 | RAB1A | 2370671 | 3235 | 0.249 | 0.4811 | Yes | ||
| 45 | CACNB2 | 1500095 7330707 | 3340 | 0.234 | 0.4790 | Yes | ||
| 46 | SPRED1 | 6940706 | 3357 | 0.231 | 0.4816 | Yes | ||
| 47 | STC1 | 360161 | 3377 | 0.228 | 0.4840 | Yes | ||
| 48 | RGS1 | 4060347 4540181 | 3385 | 0.228 | 0.4870 | Yes | ||
| 49 | DYRK1A | 3190181 | 3402 | 0.226 | 0.4895 | Yes | ||
| 50 | EED | 2320373 | 3557 | 0.204 | 0.4842 | Yes | ||
| 51 | MARK1 | 450484 | 3618 | 0.198 | 0.4839 | Yes | ||
| 52 | RIN2 | 2450184 | 3643 | 0.194 | 0.4855 | Yes | ||
| 53 | SOX11 | 610279 | 3649 | 0.194 | 0.4881 | Yes | ||
| 54 | FGA | 4210056 5220239 | 3653 | 0.193 | 0.4909 | Yes | ||
| 55 | DDEF1 | 1170411 4070465 | 3659 | 0.193 | 0.4935 | Yes | ||
| 56 | MMP15 | 5900717 | 3674 | 0.192 | 0.4956 | Yes | ||
| 57 | RASD2 | 3360010 | 3759 | 0.181 | 0.4937 | No | ||
| 58 | SOX9 | 5720681 | 3838 | 0.172 | 0.4921 | No | ||
| 59 | SLC1A1 | 520372 6420059 | 3915 | 0.165 | 0.4904 | No | ||
| 60 | FOS | 1850315 | 3972 | 0.161 | 0.4898 | No | ||
| 61 | PLCG1 | 6020369 | 3983 | 0.160 | 0.4916 | No | ||
| 62 | HAS2 | 5360181 | 4177 | 0.142 | 0.4833 | No | ||
| 63 | GPR85 | 5130750 5910020 | 4414 | 0.126 | 0.4724 | No | ||
| 64 | ABCC5 | 2100600 5050692 | 4434 | 0.124 | 0.4732 | No | ||
| 65 | DLG5 | 450215 | 4711 | 0.107 | 0.4598 | No | ||
| 66 | ROBO2 | 450136 | 4745 | 0.105 | 0.4596 | No | ||
| 67 | SRPK2 | 6380341 | 4887 | 0.098 | 0.4534 | No | ||
| 68 | PRKAA1 | 510156 | 4900 | 0.098 | 0.4542 | No | ||
| 69 | USP38 | 4590095 | 4915 | 0.097 | 0.4549 | No | ||
| 70 | NUDT11 | 3840239 | 4917 | 0.097 | 0.4563 | No | ||
| 71 | PDE4A | 1190619 1340129 5720100 6370035 | 4993 | 0.094 | 0.4537 | No | ||
| 72 | ANKRD17 | 2120402 3850035 6400044 | 5390 | 0.077 | 0.4333 | No | ||
| 73 | MBNL1 | 2640762 7100048 | 5455 | 0.075 | 0.4310 | No | ||
| 74 | ZCCHC2 | 460592 | 5478 | 0.075 | 0.4309 | No | ||
| 75 | CAV3 | 1770519 | 5794 | 0.064 | 0.4148 | No | ||
| 76 | LASS2 | 4120086 6770358 | 5881 | 0.062 | 0.4111 | No | ||
| 77 | PRPF4B | 940181 2570300 | 5922 | 0.061 | 0.4098 | No | ||
| 78 | SMARCA1 | 4230315 | 6029 | 0.058 | 0.4049 | No | ||
| 79 | PRKCE | 5700053 | 6158 | 0.054 | 0.3988 | No | ||
| 80 | TRIB1 | 2320435 | 6344 | 0.050 | 0.3895 | No | ||
| 81 | COL10A1 | 2360307 | 6426 | 0.048 | 0.3858 | No | ||
| 82 | KCNH7 | 7510068 | 6444 | 0.047 | 0.3856 | No | ||
| 83 | RAB5A | 6520324 | 6458 | 0.047 | 0.3856 | No | ||
| 84 | LRRN3 | 1450133 | 6510 | 0.046 | 0.3835 | No | ||
| 85 | RNF38 | 1940746 3140133 3840601 | 6777 | 0.041 | 0.3697 | No | ||
| 86 | FEM1C | 60300 6130717 6980164 | 6929 | 0.037 | 0.3621 | No | ||
| 87 | ASPN | 2450021 | 7008 | 0.036 | 0.3584 | No | ||
| 88 | HOXD4 | 5900301 | 7297 | 0.031 | 0.3432 | No | ||
| 89 | ACCN2 | 6450465 | 7301 | 0.031 | 0.3435 | No | ||
| 90 | DDIT4 | 2570408 | 7463 | 0.028 | 0.3352 | No | ||
| 91 | PPFIA4 | 630446 | 7577 | 0.026 | 0.3294 | No | ||
| 92 | NDFIP1 | 1980402 | 7640 | 0.025 | 0.3265 | No | ||
| 93 | USP47 | 2640497 | 7789 | 0.023 | 0.3188 | No | ||
| 94 | MYRIP | 1580471 | 7796 | 0.023 | 0.3188 | No | ||
| 95 | NLK | 2030010 2450041 | 7808 | 0.022 | 0.3185 | No | ||
| 96 | UBE2D1 | 1850400 | 7951 | 0.020 | 0.3111 | No | ||
| 97 | FLRT3 | 70685 2760497 6040519 | 8209 | 0.016 | 0.2974 | No | ||
| 98 | HNRPF | 670138 | 8367 | 0.014 | 0.2891 | No | ||
| 99 | CLCN3 | 510195 780176 1660093 3830372 | 8419 | 0.013 | 0.2866 | No | ||
| 100 | PACRG | 3140053 | 8458 | 0.013 | 0.2847 | No | ||
| 101 | CTTNBP2 | 2650167 | 8538 | 0.012 | 0.2806 | No | ||
| 102 | CDH11 | 1230133 2100180 4610110 | 8670 | 0.010 | 0.2736 | No | ||
| 103 | DR1 | 5860750 | 8689 | 0.009 | 0.2728 | No | ||
| 104 | KHDRBS3 | 2120154 | 8713 | 0.009 | 0.2717 | No | ||
| 105 | TLK2 | 1170161 | 8891 | 0.007 | 0.2622 | No | ||
| 106 | DNMT3A | 4050068 4810717 | 8894 | 0.006 | 0.2621 | No | ||
| 107 | POGZ | 870348 7040286 | 8907 | 0.006 | 0.2616 | No | ||
| 108 | FZD6 | 360288 | 9067 | 0.004 | 0.2530 | No | ||
| 109 | IGFBP5 | 2360592 | 9180 | 0.003 | 0.2470 | No | ||
| 110 | TMEFF1 | 2680100 | 9259 | 0.001 | 0.2428 | No | ||
| 111 | CPEB3 | 3940164 | 9319 | 0.000 | 0.2396 | No | ||
| 112 | FZD4 | 1580520 | 9457 | -0.001 | 0.2322 | No | ||
| 113 | KIF1A | 450632 | 9626 | -0.003 | 0.2231 | No | ||
| 114 | TSC22D1 | 1340739 6040181 | 9654 | -0.004 | 0.2217 | No | ||
| 115 | PCDH8 | 2360047 4200452 | 9749 | -0.005 | 0.2167 | No | ||
| 116 | PTGS2 | 2510301 3170369 | 9769 | -0.005 | 0.2157 | No | ||
| 117 | EYA1 | 1450278 5220390 | 9836 | -0.006 | 0.2122 | No | ||
| 118 | PUM2 | 4200441 5910446 | 10122 | -0.011 | 0.1969 | No | ||
| 119 | BCL9 | 7100112 | 10228 | -0.012 | 0.1914 | No | ||
| 120 | BICD2 | 1090411 | 10554 | -0.017 | 0.1740 | No | ||
| 121 | FBXW11 | 6450632 | 10657 | -0.018 | 0.1688 | No | ||
| 122 | CCNJ | 870047 | 10929 | -0.023 | 0.1544 | No | ||
| 123 | TEAD3 | 2320142 | 11693 | -0.037 | 0.1136 | No | ||
| 124 | NRK | 4590609 | 11782 | -0.038 | 0.1094 | No | ||
| 125 | DDX3Y | 1580278 4200519 | 11964 | -0.042 | 0.1002 | No | ||
| 126 | THRB | 1780673 | 11988 | -0.043 | 0.0996 | No | ||
| 127 | EXOC5 | 7000711 | 12156 | -0.046 | 0.0912 | No | ||
| 128 | MAGI1 | 6510500 1990239 4670471 | 12343 | -0.051 | 0.0819 | No | ||
| 129 | FLRT2 | 4200035 | 12363 | -0.051 | 0.0816 | No | ||
| 130 | LRRC4 | 3850041 | 12468 | -0.054 | 0.0768 | No | ||
| 131 | AEBP2 | 2970603 | 12684 | -0.060 | 0.0660 | No | ||
| 132 | SIX4 | 6760022 | 12830 | -0.064 | 0.0591 | No | ||
| 133 | CDYL | 4200142 4730397 6060400 | 12958 | -0.069 | 0.0533 | No | ||
| 134 | QKI | 5220093 6130044 | 13074 | -0.073 | 0.0481 | No | ||
| 135 | ADAMTS10 | 730563 | 13111 | -0.074 | 0.0473 | No | ||
| 136 | RIPK5 | 1230647 1340114 | 13357 | -0.084 | 0.0352 | No | ||
| 137 | ZFHX4 | 6110167 | 13375 | -0.085 | 0.0356 | No | ||
| 138 | NEGR1 | 1940731 5130170 | 13465 | -0.088 | 0.0321 | No | ||
| 139 | PALM2 | 4120068 7100398 | 13540 | -0.092 | 0.0294 | No | ||
| 140 | TAL1 | 7040239 | 13685 | -0.098 | 0.0231 | No | ||
| 141 | SOCS5 | 3830398 7100093 | 13768 | -0.103 | 0.0202 | No | ||
| 142 | RBBP7 | 430113 450450 2370309 | 13886 | -0.110 | 0.0155 | No | ||
| 143 | BTBD3 | 1980041 2510577 3190053 4200746 | 13908 | -0.111 | 0.0160 | No | ||
| 144 | LRRN1 | 3290154 | 14027 | -0.117 | 0.0114 | No | ||
| 145 | TGFBR1 | 1400148 4280020 6550711 | 14036 | -0.118 | 0.0127 | No | ||
| 146 | ING3 | 110438 2060390 | 14093 | -0.121 | 0.0115 | No | ||
| 147 | DCBLD2 | 5890333 | 14096 | -0.121 | 0.0132 | No | ||
| 148 | PDE4D | 2470528 6660014 | 14192 | -0.128 | 0.0099 | No | ||
| 149 | REV3L | 1090717 | 14218 | -0.130 | 0.0105 | No | ||
| 150 | ATRX | 2340039 3610132 4210373 | 14242 | -0.131 | 0.0112 | No | ||
| 151 | NEUROD1 | 3060619 | 14437 | -0.145 | 0.0029 | No | ||
| 152 | DAG1 | 460053 610341 | 14568 | -0.157 | -0.0018 | No | ||
| 153 | MAK | 2940706 | 14645 | -0.164 | -0.0035 | No | ||
| 154 | RAPH1 | 6760411 | 14720 | -0.170 | -0.0050 | No | ||
| 155 | LRRFIP2 | 670575 730152 6590438 | 14766 | -0.175 | -0.0048 | No | ||
| 156 | GLRA2 | 7000019 | 14829 | -0.181 | -0.0054 | No | ||
| 157 | PANK3 | 3060687 | 14840 | -0.182 | -0.0033 | No | ||
| 158 | FBXW7 | 4210338 7050280 | 14925 | -0.190 | -0.0050 | No | ||
| 159 | CDH5 | 5340487 | 15220 | -0.225 | -0.0176 | No | ||
| 160 | LMNB1 | 5890292 6020008 | 15275 | -0.233 | -0.0170 | No | ||
| 161 | STAG2 | 4540132 | 15364 | -0.245 | -0.0181 | No | ||
| 162 | ADCY6 | 450364 6290670 6940286 | 15376 | -0.247 | -0.0150 | No | ||
| 163 | PURG | 1190593 | 15637 | -0.291 | -0.0248 | No | ||
| 164 | MPPE1 | 5080364 7050739 | 15645 | -0.293 | -0.0208 | No | ||
| 165 | CDK5R1 | 3870161 | 15770 | -0.314 | -0.0228 | No | ||
| 166 | PREI3 | 3170066 3440075 6370427 | 15818 | -0.324 | -0.0205 | No | ||
| 167 | SLC39A10 | 4150347 | 15906 | -0.341 | -0.0201 | No | ||
| 168 | TXNDC4 | 450537 | 15958 | -0.352 | -0.0176 | No | ||
| 169 | MYST2 | 4540494 | 16154 | -0.398 | -0.0222 | No | ||
| 170 | PAPOLG | 2470400 | 16216 | -0.413 | -0.0194 | No | ||
| 171 | ERBB2IP | 580253 1090672 | 16238 | -0.419 | -0.0143 | No | ||
| 172 | BZW2 | 940079 | 16510 | -0.495 | -0.0216 | No | ||
| 173 | UBE2A | 4010563 | 16569 | -0.516 | -0.0170 | No | ||
| 174 | UBE2D3 | 3190452 | 16628 | -0.536 | -0.0121 | No | ||
| 175 | ARID1A | 2630022 1690551 4810110 | 16639 | -0.539 | -0.0046 | No | ||
| 176 | PPFIA1 | 430176 3610524 | 16660 | -0.546 | 0.0025 | No | ||
| 177 | TULP4 | 2320364 | 16788 | -0.586 | 0.0044 | No | ||
| 178 | SMARCD1 | 3060193 3850184 6400369 | 17317 | -0.794 | -0.0124 | No | ||
| 179 | TERF2 | 3840044 | 17401 | -0.837 | -0.0044 | No | ||
| 180 | BAZ2A | 730184 | 17764 | -1.024 | -0.0087 | No | ||
| 181 | JUNB | 4230048 | 17821 | -1.073 | 0.0043 | No | ||
| 182 | OGT | 2360131 4610333 | 18039 | -1.238 | 0.0110 | No | ||
| 183 | UBE2Q1 | 610348 2350114 4210717 | 18147 | -1.351 | 0.0254 | No |