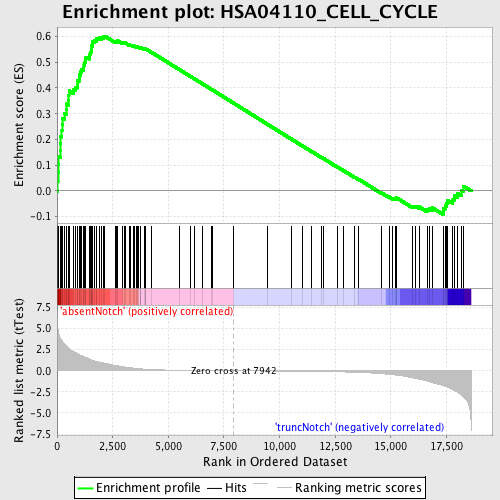
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | HSA04110_CELL_CYCLE |
| Enrichment Score (ES) | 0.60140747 |
| Normalized Enrichment Score (NES) | 1.7376692 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.034962736 |
| FWER p-Value | 0.385 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HDAC2 | 4050433 | 20 | 5.359 | 0.0374 | Yes | ||
| 2 | ANAPC1 | 6350162 | 41 | 4.838 | 0.0710 | Yes | ||
| 3 | YWHAB | 1740176 | 56 | 4.653 | 0.1036 | Yes | ||
| 4 | SMAD4 | 5670519 | 72 | 4.465 | 0.1348 | Yes | ||
| 5 | ANAPC4 | 4540338 | 147 | 3.854 | 0.1585 | Yes | ||
| 6 | RBL1 | 3130372 | 151 | 3.829 | 0.1858 | Yes | ||
| 7 | ORC6L | 2260592 | 165 | 3.774 | 0.2121 | Yes | ||
| 8 | CCNH | 3190347 | 215 | 3.563 | 0.2350 | Yes | ||
| 9 | MCM4 | 2760673 5420711 | 225 | 3.505 | 0.2597 | Yes | ||
| 10 | SKP2 | 360711 380093 4810368 | 246 | 3.435 | 0.2832 | Yes | ||
| 11 | PCNA | 940754 | 337 | 3.090 | 0.3005 | Yes | ||
| 12 | RB1 | 5900338 | 411 | 2.912 | 0.3175 | Yes | ||
| 13 | MAD2L1 | 4480725 | 417 | 2.886 | 0.3379 | Yes | ||
| 14 | GSK3B | 5360348 | 510 | 2.696 | 0.3523 | Yes | ||
| 15 | CCNE2 | 3120537 | 529 | 2.639 | 0.3702 | Yes | ||
| 16 | CREBBP | 5690035 7040050 | 535 | 2.625 | 0.3888 | Yes | ||
| 17 | ORC5L | 1940133 1940711 | 754 | 2.234 | 0.3930 | Yes | ||
| 18 | CCNB1 | 4590433 4780372 | 841 | 2.121 | 0.4036 | Yes | ||
| 19 | SMAD2 | 4200592 | 912 | 2.023 | 0.4143 | Yes | ||
| 20 | ANAPC5 | 730164 7100685 | 922 | 2.010 | 0.4283 | Yes | ||
| 21 | TFDP1 | 1980112 | 986 | 1.906 | 0.4385 | Yes | ||
| 22 | CDKN1C | 6520577 | 996 | 1.891 | 0.4516 | Yes | ||
| 23 | BUB1B | 1450288 | 1041 | 1.842 | 0.4625 | Yes | ||
| 24 | HDAC1 | 2850670 | 1077 | 1.803 | 0.4735 | Yes | ||
| 25 | CDC25A | 3800184 | 1196 | 1.670 | 0.4791 | Yes | ||
| 26 | YWHAQ | 6760524 | 1207 | 1.661 | 0.4905 | Yes | ||
| 27 | CUL1 | 1990632 | 1241 | 1.620 | 0.5003 | Yes | ||
| 28 | MDM2 | 3450053 5080138 | 1273 | 1.581 | 0.5100 | Yes | ||
| 29 | CDC16 | 1940706 | 1295 | 1.556 | 0.5200 | Yes | ||
| 30 | CDKN2A | 4670215 4760047 | 1438 | 1.404 | 0.5224 | Yes | ||
| 31 | ATR | 6860273 | 1459 | 1.381 | 0.5312 | Yes | ||
| 32 | CCND3 | 380528 730500 | 1491 | 1.356 | 0.5393 | Yes | ||
| 33 | MCM6 | 60092 540181 6510110 | 1526 | 1.312 | 0.5468 | Yes | ||
| 34 | E2F1 | 5360093 | 1556 | 1.284 | 0.5545 | Yes | ||
| 35 | YWHAZ | 1230717 | 1558 | 1.283 | 0.5636 | Yes | ||
| 36 | YWHAG | 3780341 | 1591 | 1.251 | 0.5709 | Yes | ||
| 37 | CCND2 | 5340167 | 1608 | 1.231 | 0.5789 | Yes | ||
| 38 | CCNE1 | 110064 | 1670 | 1.172 | 0.5840 | Yes | ||
| 39 | GADD45G | 2510142 | 1766 | 1.097 | 0.5867 | Yes | ||
| 40 | CDK2 | 130484 2260301 4010088 5050110 | 1778 | 1.088 | 0.5939 | Yes | ||
| 41 | CDC7 | 4060546 4850041 | 1905 | 1.007 | 0.5943 | Yes | ||
| 42 | RBX1 | 2120010 2340047 | 1998 | 0.965 | 0.5963 | Yes | ||
| 43 | YWHAE | 5310435 | 2086 | 0.911 | 0.5981 | Yes | ||
| 44 | CDC14A | 4050132 | 2143 | 0.881 | 0.6014 | Yes | ||
| 45 | MCM3 | 5570068 | 2610 | 0.627 | 0.5807 | No | ||
| 46 | MCM7 | 3290292 5220056 | 2662 | 0.607 | 0.5823 | No | ||
| 47 | CDC6 | 4570296 5360600 | 2713 | 0.578 | 0.5838 | No | ||
| 48 | E2F2 | 5270609 5570377 7000465 | 2918 | 0.493 | 0.5763 | No | ||
| 49 | ORC3L | 3060348 | 3016 | 0.456 | 0.5743 | No | ||
| 50 | CCNA2 | 5290075 | 3056 | 0.440 | 0.5754 | No | ||
| 51 | MAD1L1 | 5700035 | 3238 | 0.379 | 0.5683 | No | ||
| 52 | E2F3 | 50162 460180 | 3320 | 0.350 | 0.5664 | No | ||
| 53 | WEE1 | 3390070 | 3430 | 0.317 | 0.5628 | No | ||
| 54 | CDC45L | 70537 3130114 | 3466 | 0.305 | 0.5631 | No | ||
| 55 | GADD45B | 2350408 | 3576 | 0.272 | 0.5592 | No | ||
| 56 | RBL2 | 580446 1400670 | 3592 | 0.267 | 0.5603 | No | ||
| 57 | YWHAH | 1660133 2810053 | 3653 | 0.251 | 0.5589 | No | ||
| 58 | CDKN1A | 4050088 6400706 | 3730 | 0.231 | 0.5564 | No | ||
| 59 | ORC2L | 1990470 6510019 | 3731 | 0.231 | 0.5581 | No | ||
| 60 | CDK6 | 4920253 | 3755 | 0.226 | 0.5584 | No | ||
| 61 | CHEK1 | 50167 1940300 | 3941 | 0.188 | 0.5498 | No | ||
| 62 | ABL1 | 1050593 2030050 4010114 | 3953 | 0.186 | 0.5505 | No | ||
| 63 | CDC23 | 3190593 | 3954 | 0.186 | 0.5519 | No | ||
| 64 | ORC4L | 4230538 5550288 | 3986 | 0.180 | 0.5515 | No | ||
| 65 | CDKN2B | 6020040 | 4239 | 0.141 | 0.5389 | No | ||
| 66 | CDC14B | 2320497 3360162 | 5503 | 0.055 | 0.4711 | No | ||
| 67 | CDC25C | 2570673 4760161 6520707 | 6003 | 0.040 | 0.4444 | No | ||
| 68 | TGFB2 | 4920292 | 6193 | 0.034 | 0.4344 | No | ||
| 69 | SMC1B | 2450397 | 6515 | 0.027 | 0.4173 | No | ||
| 70 | TGFB3 | 1070041 | 6952 | 0.017 | 0.3939 | No | ||
| 71 | CCNB2 | 6510528 | 6991 | 0.017 | 0.3919 | No | ||
| 72 | CCNA1 | 6290113 | 7918 | 0.000 | 0.3419 | No | ||
| 73 | PRKDC | 1400072 4230541 | 9467 | -0.026 | 0.2585 | No | ||
| 74 | ATM | 3610110 4050524 | 10515 | -0.048 | 0.2023 | No | ||
| 75 | CDK7 | 2640451 | 11047 | -0.060 | 0.1740 | No | ||
| 76 | ANAPC10 | 870086 1170037 2260129 | 11439 | -0.072 | 0.1534 | No | ||
| 77 | SFN | 6290301 7510608 | 11872 | -0.087 | 0.1307 | No | ||
| 78 | MCM2 | 5050139 | 11994 | -0.092 | 0.1248 | No | ||
| 79 | CCNB3 | 3360600 | 12623 | -0.124 | 0.0918 | No | ||
| 80 | PLK1 | 1780369 2640121 | 12890 | -0.143 | 0.0785 | No | ||
| 81 | ANAPC11 | 1780601 | 13373 | -0.179 | 0.0537 | No | ||
| 82 | BUB1 | 5390270 | 13543 | -0.197 | 0.0460 | No | ||
| 83 | GADD45A | 2900717 | 14582 | -0.346 | -0.0076 | No | ||
| 84 | CCND1 | 460524 770309 3120576 6980398 | 14927 | -0.419 | -0.0232 | No | ||
| 85 | CDKN2C | 5050750 5130148 | 15096 | -0.464 | -0.0289 | No | ||
| 86 | TGFB1 | 1940162 | 15204 | -0.500 | -0.0311 | No | ||
| 87 | CDKN1B | 3800025 6450044 | 15233 | -0.512 | -0.0289 | No | ||
| 88 | CHEK2 | 610139 1050022 | 15235 | -0.512 | -0.0253 | No | ||
| 89 | MCM5 | 2680647 | 15984 | -0.836 | -0.0597 | No | ||
| 90 | CDC26 | 5570279 | 16103 | -0.902 | -0.0596 | No | ||
| 91 | SMC1A | 3060600 5700148 5890113 6370154 | 16276 | -1.001 | -0.0617 | No | ||
| 92 | ORC1L | 2370328 6110390 | 16632 | -1.219 | -0.0722 | No | ||
| 93 | FZR1 | 6220504 6660670 | 16735 | -1.294 | -0.0684 | No | ||
| 94 | CDKN2D | 6040035 | 16870 | -1.413 | -0.0655 | No | ||
| 95 | SMAD3 | 6450671 | 17349 | -1.764 | -0.0787 | No | ||
| 96 | ANAPC2 | 6660438 | 17382 | -1.790 | -0.0676 | No | ||
| 97 | TP53 | 6130707 | 17437 | -1.852 | -0.0572 | No | ||
| 98 | PTTG1 | 520463 | 17502 | -1.903 | -0.0470 | No | ||
| 99 | CDK4 | 540075 4540600 | 17551 | -1.953 | -0.0356 | No | ||
| 100 | MAD2L2 | 1240358 | 17792 | -2.237 | -0.0325 | No | ||
| 101 | CDC20 | 3440017 3440044 6220088 | 17846 | -2.302 | -0.0189 | No | ||
| 102 | BUB3 | 3170546 | 17989 | -2.521 | -0.0085 | No | ||
| 103 | ANAPC7 | 5260274 | 18170 | -2.815 | 0.0020 | No | ||
| 104 | CDC25B | 6940102 | 18270 | -3.071 | 0.0187 | No |

