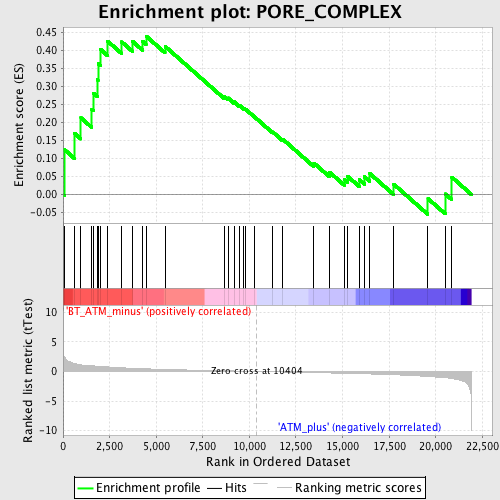
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | PORE_COMPLEX |
| Enrichment Score (ES) | 0.43961757 |
| Normalized Enrichment Score (NES) | 1.4806852 |
| Nominal p-value | 0.045558088 |
| FDR q-value | 0.9719443 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NUTF2 | 1434367_s_at 1447440_at 1449017_at | 65 | 2.441 | 0.1251 | Yes | ||
| 2 | NUP153 | 1441689_at 1441733_s_at 1452176_at | 616 | 1.331 | 0.1698 | Yes | ||
| 3 | KPNA1 | 1419548_at 1449503_at 1449504_at 1460260_s_at | 957 | 1.116 | 0.2128 | Yes | ||
| 4 | RANBP2 | 1422621_at 1440104_at 1445883_at 1450690_at | 1542 | 0.960 | 0.2365 | Yes | ||
| 5 | KPNA3 | 1421828_at 1445527_at 1446538_at 1450385_at 1450386_at | 1638 | 0.939 | 0.2815 | Yes | ||
| 6 | NUP214 | 1434351_at 1456503_at 1457815_at | 1832 | 0.894 | 0.3196 | Yes | ||
| 7 | HRB | 1426922_s_at 1426923_at 1434840_at 1452237_at 1452238_at | 1878 | 0.885 | 0.3639 | Yes | ||
| 8 | NUP98 | 1434220_at 1439154_at 1446397_at 1446988_at | 2006 | 0.854 | 0.4029 | Yes | ||
| 9 | DDX19B | 1424590_at 1425203_at 1425205_at 1442444_at | 2385 | 0.767 | 0.4259 | Yes | ||
| 10 | NUP50 | 1422719_s_at 1428328_at 1450722_at | 3150 | 0.620 | 0.4235 | Yes | ||
| 11 | NUP160 | 1418530_at 1444892_at | 3723 | 0.532 | 0.4253 | Yes | ||
| 12 | BAX | 1416837_at | 4273 | 0.465 | 0.4246 | Yes | ||
| 13 | NUP62 | 1415925_a_at 1415926_at 1437612_at 1438917_x_at 1447905_x_at 1448150_at | 4456 | 0.444 | 0.4396 | Yes | ||
| 14 | AAAS | 1445494_at 1451377_a_at | 5473 | 0.347 | 0.4114 | No | ||
| 15 | VDAC1 | 1415998_at 1436992_x_at 1437192_x_at 1437452_x_at 1437947_x_at | 8653 | 0.110 | 0.2720 | No | ||
| 16 | NXT1 | 1422488_at | 8855 | 0.099 | 0.2680 | No | ||
| 17 | TMEM48 | 1424173_at 1433813_at 1460353_at | 9181 | 0.078 | 0.2573 | No | ||
| 18 | EIF5A | 1437859_x_at 1451470_s_at | 9451 | 0.061 | 0.2482 | No | ||
| 19 | TPR | 1426948_at 1426949_s_at 1456112_at 1456651_a_at | 9697 | 0.045 | 0.2394 | No | ||
| 20 | C8A | 1428012_at | 9788 | 0.040 | 0.2374 | No | ||
| 21 | SUMO1 | 1438289_a_at 1444557_at 1451005_at 1456349_x_at | 10266 | 0.009 | 0.2161 | No | ||
| 22 | IPO7 | 1442511_at 1454955_at 1458165_at | 11255 | -0.054 | 0.1738 | No | ||
| 23 | NUP88 | 1428073_a_at | 11802 | -0.086 | 0.1534 | No | ||
| 24 | SENP2 | 1425465_a_at 1425466_at 1451717_s_at 1455327_at | 13466 | -0.192 | 0.0876 | No | ||
| 25 | NUP54 | 1432539_a_at 1433580_at 1438951_x_at 1447253_x_at 1459651_s_at | 14302 | -0.248 | 0.0625 | No | ||
| 26 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 15097 | -0.307 | 0.0423 | No | ||
| 27 | NUP107 | 1426751_s_at | 15269 | -0.320 | 0.0513 | No | ||
| 28 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 15903 | -0.369 | 0.0417 | No | ||
| 29 | RAE1 | 1429528_at 1444256_at 1459822_at | 16187 | -0.393 | 0.0494 | No | ||
| 30 | BAK1 | 1418991_at 1425716_s_at | 16471 | -0.419 | 0.0585 | No | ||
| 31 | NUP133 | 1423787_at 1423788_at 1436816_at 1441235_at 1451111_at 1457542_at | 17756 | -0.551 | 0.0287 | No | ||
| 32 | SNUPN | 1441942_x_at 1445434_at 1452808_at | 19593 | -0.841 | -0.0110 | No | ||
| 33 | RANBP5 | 1426945_at 1426946_at 1431706_at | 20525 | -1.055 | 0.0019 | No | ||
| 34 | XPO7 | 1415682_at 1435814_at 1439411_a_at 1442836_at 1446826_at | 20876 | -1.190 | 0.0483 | No |

