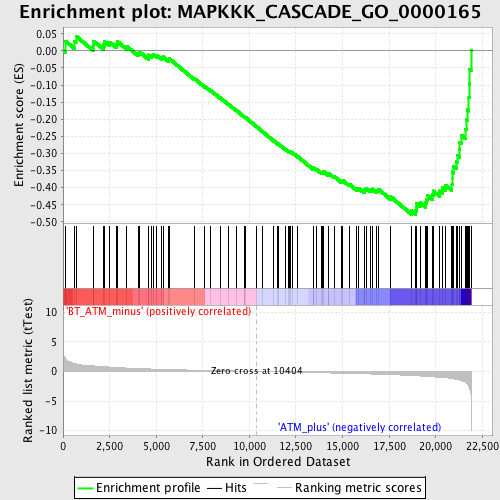
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | MAPKKK_CASCADE_GO_0000165 |
| Enrichment Score (ES) | -0.4784316 |
| Normalized Enrichment Score (NES) | -1.8488423 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.116214246 |
| FWER p-Value | 0.637 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 122 | 2.239 | 0.0288 | No | ||
| 2 | MAP2K4 | 1426233_at 1442679_at 1451982_at | 597 | 1.346 | 0.0278 | No | ||
| 3 | SOD1 | 1435304_at 1440222_at 1440896_at 1451124_at 1459976_s_at | 693 | 1.278 | 0.0431 | No | ||
| 4 | GRM4 | 1457299_at | 1619 | 0.943 | 0.0153 | No | ||
| 5 | MDFIC | 1427040_at | 1641 | 0.938 | 0.0287 | No | ||
| 6 | MBIP | 1424405_at | 2163 | 0.818 | 0.0175 | No | ||
| 7 | DUSP10 | 1417163_at 1417164_at | 2241 | 0.799 | 0.0262 | No | ||
| 8 | ADORA2B | 1434430_s_at 1434431_x_at 1434772_at 1450214_at | 2503 | 0.741 | 0.0257 | No | ||
| 9 | AMBP | 1416649_at | 2858 | 0.672 | 0.0198 | No | ||
| 10 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 2920 | 0.662 | 0.0272 | No | ||
| 11 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 3414 | 0.576 | 0.0135 | No | ||
| 12 | SPRED2 | 1423001_at 1434403_at 1436892_at 1441415_at 1441802_at | 4023 | 0.496 | -0.0068 | No | ||
| 13 | MAP3K10 | 1436373_at 1440500_at 1457456_at 1458651_at | 4116 | 0.482 | -0.0036 | No | ||
| 14 | MAP4K3 | 1444406_at | 4587 | 0.431 | -0.0184 | No | ||
| 15 | MAP3K9 | 1432981_at 1442218_at 1455684_at 1458922_at | 4593 | 0.431 | -0.0120 | No | ||
| 16 | MAP3K2 | 1422250_at | 4767 | 0.414 | -0.0136 | No | ||
| 17 | MAP4K5 | 1427083_a_at 1427084_a_at 1427376_a_at 1440059_at 1445426_at 1446348_at 1447204_at | 4836 | 0.406 | -0.0105 | No | ||
| 18 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 5018 | 0.389 | -0.0128 | No | ||
| 19 | MAPK8IP1 | 1425679_a_at 1440619_at | 5286 | 0.364 | -0.0194 | No | ||
| 20 | FGFR3 | 1421841_at 1425796_a_at | 5364 | 0.356 | -0.0175 | No | ||
| 21 | MAPK9 | 1421876_at 1421877_at 1421878_at | 5667 | 0.330 | -0.0262 | No | ||
| 22 | TNIK | 1443802_at | 5691 | 0.327 | -0.0222 | No | ||
| 23 | FGF13 | 1418497_at 1418498_at | 7036 | 0.220 | -0.0804 | No | ||
| 24 | SHC1 | 1422853_at 1422854_at | 7599 | 0.182 | -0.1033 | No | ||
| 25 | DUSP8 | 1418714_at | 7924 | 0.159 | -0.1157 | No | ||
| 26 | MAP3K6 | 1449901_a_at | 8442 | 0.124 | -0.1375 | No | ||
| 27 | CHRNA7 | 1440681_at 1450299_at | 8872 | 0.098 | -0.1556 | No | ||
| 28 | TRAF7 | 1424320_a_at | 9315 | 0.069 | -0.1748 | No | ||
| 29 | NRTN | 1449281_at | 9729 | 0.043 | -0.1930 | No | ||
| 30 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 9747 | 0.042 | -0.1932 | No | ||
| 31 | FPR1 | 1450808_at | 9786 | 0.040 | -0.1943 | No | ||
| 32 | ADRB3 | 1421555_at 1455918_at | 10381 | 0.001 | -0.2215 | No | ||
| 33 | EDG5 | 1428176_at 1447880_x_at | 10692 | -0.017 | -0.2354 | No | ||
| 34 | FGFR1 | 1424050_s_at 1425911_a_at 1436551_at | 11314 | -0.057 | -0.2630 | No | ||
| 35 | FGF2 | 1449826_a_at | 11530 | -0.070 | -0.2717 | No | ||
| 36 | EGF | 1418093_a_at | 11572 | -0.072 | -0.2725 | No | ||
| 37 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 11953 | -0.096 | -0.2884 | No | ||
| 38 | CCM2 | 1434648_a_at 1434649_at 1456290_x_at 1457339_at | 12115 | -0.105 | -0.2942 | No | ||
| 39 | GNG3 | 1417428_at | 12143 | -0.107 | -0.2938 | No | ||
| 40 | PKN1 | 1458674_at 1459231_at | 12213 | -0.112 | -0.2952 | No | ||
| 41 | PROK2 | 1419681_a_at 1426176_a_at 1451952_at | 12309 | -0.118 | -0.2977 | No | ||
| 42 | SCG2 | 1450708_at | 12590 | -0.135 | -0.3085 | No | ||
| 43 | MAP3K11 | 1450669_at | 13441 | -0.190 | -0.3445 | No | ||
| 44 | PIK3CB | 1445731_at 1453069_at | 13458 | -0.191 | -0.3423 | No | ||
| 45 | MAP3K12 | 1417115_at 1438908_at 1456565_s_at | 13607 | -0.201 | -0.3460 | No | ||
| 46 | GPS1 | 1415699_a_at | 13867 | -0.219 | -0.3544 | No | ||
| 47 | MAPK10 | 1437195_x_at 1448342_at 1458002_at 1458062_at 1458513_at | 13942 | -0.224 | -0.3544 | No | ||
| 48 | ADRA2A | 1423022_at 1433600_at 1433601_at | 13962 | -0.226 | -0.3518 | No | ||
| 49 | GHRL | 1448980_at | 14238 | -0.244 | -0.3606 | No | ||
| 50 | TPD52L1 | 1418412_at | 14274 | -0.247 | -0.3584 | No | ||
| 51 | TGFA | 1421942_s_at 1450421_at | 14563 | -0.268 | -0.3675 | No | ||
| 52 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 14964 | -0.297 | -0.3812 | No | ||
| 53 | DAXX | 1419026_at | 15027 | -0.302 | -0.3794 | No | ||
| 54 | ATP6AP2 | 1423662_at 1437688_x_at 1439456_x_at | 15364 | -0.326 | -0.3898 | No | ||
| 55 | MDFI | 1420713_a_at 1425924_at | 15751 | -0.355 | -0.4020 | No | ||
| 56 | TDGF1 | 1450989_at | 15863 | -0.365 | -0.4015 | No | ||
| 57 | PLCE1 | 1452398_at | 16167 | -0.391 | -0.4094 | No | ||
| 58 | MAPKAPK2 | 1426648_at | 16184 | -0.392 | -0.4041 | No | ||
| 59 | C5AR1 | 1422190_at 1439902_at | 16283 | -0.400 | -0.4024 | No | ||
| 60 | TAOK2 | 1438208_at 1443642_at 1456826_at | 16503 | -0.422 | -0.4059 | No | ||
| 61 | DUSP22 | 1441723_at 1448985_at | 16601 | -0.429 | -0.4038 | No | ||
| 62 | GPS2 | 1417796_at | 16851 | -0.454 | -0.4082 | No | ||
| 63 | ADRA2C | 1422335_at 1435296_at 1439762_x_at 1457328_at | 16924 | -0.461 | -0.4044 | No | ||
| 64 | CXCR4 | 1448710_at | 17598 | -0.531 | -0.4271 | No | ||
| 65 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 18721 | -0.687 | -0.4679 | Yes | ||
| 66 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 18935 | -0.719 | -0.4666 | Yes | ||
| 67 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 18979 | -0.727 | -0.4574 | Yes | ||
| 68 | SPRED1 | 1423160_at 1423161_s_at 1423162_s_at 1428777_at 1441517_at 1442859_at 1443652_x_at 1445228_at 1452911_at 1460116_s_at | 18988 | -0.728 | -0.4465 | Yes | ||
| 69 | CD81 | 1416330_at | 19177 | -0.760 | -0.4435 | Yes | ||
| 70 | MINK1 | 1449362_a_at | 19474 | -0.816 | -0.4445 | Yes | ||
| 71 | CARTPT | 1422825_at | 19522 | -0.825 | -0.4340 | Yes | ||
| 72 | HIPK2 | 1424863_a_at 1424864_at 1425983_x_at 1428433_at 1429566_a_at 1432523_at 1446167_at 1448631_a_at 1456022_at 1458520_at | 19583 | -0.838 | -0.4238 | Yes | ||
| 73 | PTPLAD1 | 1433706_a_at 1452427_s_at 1454698_at | 19826 | -0.891 | -0.4212 | Yes | ||
| 74 | EDA2R | 1440085_at | 19878 | -0.902 | -0.4097 | Yes | ||
| 75 | RGS4 | 1416286_at 1416287_at 1448285_at | 20211 | -0.984 | -0.4097 | Yes | ||
| 76 | DBNL | 1460334_at | 20352 | -1.014 | -0.4006 | Yes | ||
| 77 | MAP3K7IP1 | 1426898_at 1447692_x_at | 20534 | -1.058 | -0.3926 | Yes | ||
| 78 | DUSP9 | 1433844_a_at 1433845_x_at 1454737_at | 20882 | -1.193 | -0.3901 | Yes | ||
| 79 | MAP3K4 | 1421450_a_at 1447667_x_at 1450253_a_at 1459800_s_at | 20890 | -1.200 | -0.3720 | Yes | ||
| 80 | TAOK3 | 1435964_a_at 1444366_at 1444881_at 1447257_at 1455733_at | 20892 | -1.200 | -0.3536 | Yes | ||
| 81 | CAMKK2 | 1424474_a_at 1424475_at 1424476_at 1455401_at | 20973 | -1.243 | -0.3382 | Yes | ||
| 82 | MAP3K3 | 1426686_s_at 1426687_at 1436522_at | 21119 | -1.317 | -0.3245 | Yes | ||
| 83 | GADD45G | 1453851_a_at | 21192 | -1.365 | -0.3069 | Yes | ||
| 84 | LAX1 | 1438687_at | 21276 | -1.443 | -0.2885 | Yes | ||
| 85 | MADD | 1425735_at 1455502_at | 21306 | -1.464 | -0.2673 | Yes | ||
| 86 | ADRB2 | 1437302_at | 21407 | -1.565 | -0.2478 | Yes | ||
| 87 | SH2D3C | 1415886_at | 21603 | -1.823 | -0.2287 | Yes | ||
| 88 | DUSP16 | 1418401_a_at 1440615_at 1440651_at | 21660 | -1.940 | -0.2015 | Yes | ||
| 89 | DUSP2 | 1450698_at | 21696 | -2.054 | -0.1715 | Yes | ||
| 90 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21794 | -2.532 | -0.1370 | Yes | ||
| 91 | DUSP4 | 1428834_at | 21811 | -2.663 | -0.0968 | Yes | ||
| 92 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 21839 | -2.911 | -0.0533 | Yes | ||
| 93 | DUSP6 | 1415834_at | 21907 | -3.749 | 0.0012 | Yes |

