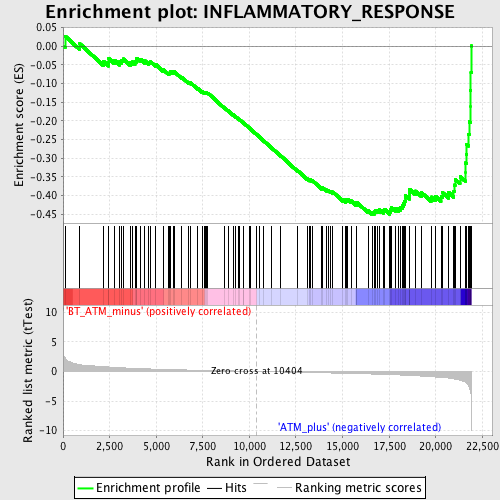
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.4505079 |
| Normalized Enrichment Score (NES) | -1.7454969 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2183408 |
| FWER p-Value | 0.963 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TGFB1 | 1420653_at 1445360_at | 106 | 2.330 | 0.0267 | No | ||
| 2 | CDO1 | 1448842_at | 897 | 1.148 | 0.0060 | No | ||
| 3 | TNFAIP6 | 1418424_at | 2151 | 0.822 | -0.0403 | No | ||
| 4 | ADORA1 | 1427331_at 1435495_at | 2439 | 0.756 | -0.0432 | No | ||
| 5 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 2454 | 0.753 | -0.0337 | No | ||
| 6 | NFATC3 | 1419976_s_at 1452497_a_at | 2750 | 0.690 | -0.0378 | No | ||
| 7 | IL5 | 1425546_a_at 1442049_at 1450550_at | 3035 | 0.639 | -0.0422 | No | ||
| 8 | CCL5 | 1418126_at | 3146 | 0.621 | -0.0388 | No | ||
| 9 | LY75 | 1449328_at | 3228 | 0.606 | -0.0343 | No | ||
| 10 | KLRG1 | 1420788_at | 3609 | 0.548 | -0.0443 | No | ||
| 11 | PLA2G7 | 1430700_a_at | 3722 | 0.532 | -0.0423 | No | ||
| 12 | FPRL1 | 1421661_at 1450583_s_at | 3862 | 0.514 | -0.0417 | No | ||
| 13 | CCL24 | 1450488_at | 3923 | 0.508 | -0.0376 | No | ||
| 14 | CXCL1 | 1419209_at 1441855_x_at 1457644_s_at | 3952 | 0.504 | -0.0320 | No | ||
| 15 | HDAC4 | 1436758_at 1447566_at 1454693_at | 4169 | 0.476 | -0.0355 | No | ||
| 16 | CEBPB | 1418901_at 1427844_a_at | 4394 | 0.452 | -0.0396 | No | ||
| 17 | ANXA1 | 1448213_at | 4607 | 0.429 | -0.0435 | No | ||
| 18 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 4701 | 0.420 | -0.0421 | No | ||
| 19 | AOC3 | 1449396_at | 4968 | 0.394 | -0.0490 | No | ||
| 20 | CCL4 | 1421578_at | 5373 | 0.355 | -0.0627 | No | ||
| 21 | AGER | 1420428_at 1441958_s_at | 5683 | 0.328 | -0.0724 | No | ||
| 22 | IL8RB | 1421734_at | 5737 | 0.323 | -0.0704 | No | ||
| 23 | CCR2 | 1421186_at 1421187_at 1421188_at 1460067_at | 5776 | 0.320 | -0.0678 | No | ||
| 24 | AOX1 | 1419435_at | 5912 | 0.307 | -0.0699 | No | ||
| 25 | GHSR | 1446756_at | 5970 | 0.302 | -0.0684 | No | ||
| 26 | IL9 | 1450565_at | 6368 | 0.271 | -0.0829 | No | ||
| 27 | KNG1 | 1416676_at 1426045_at | 6729 | 0.244 | -0.0961 | No | ||
| 28 | MEFV | 1460283_at | 6847 | 0.235 | -0.0983 | No | ||
| 29 | PLA2G2E | 1420674_at | 7222 | 0.207 | -0.1126 | No | ||
| 30 | ADORA2A | 1427519_at 1460710_at | 7480 | 0.190 | -0.1218 | No | ||
| 31 | HRH1 | 1419210_at 1438494_at | 7578 | 0.183 | -0.1238 | No | ||
| 32 | CCL3 | 1419561_at | 7670 | 0.176 | -0.1256 | No | ||
| 33 | CYBB | 1422978_at 1436778_at 1436779_at | 7703 | 0.174 | -0.1247 | No | ||
| 34 | CFHR1 | 1419436_at | 7776 | 0.170 | -0.1257 | No | ||
| 35 | EDG3 | 1437173_at 1438658_a_at 1460661_at | 8680 | 0.108 | -0.1656 | No | ||
| 36 | CHRNA7 | 1440681_at 1450299_at | 8872 | 0.098 | -0.1730 | No | ||
| 37 | APCS | 1419059_at | 9128 | 0.082 | -0.1836 | No | ||
| 38 | CD97 | 1418394_a_at | 9272 | 0.072 | -0.1892 | No | ||
| 39 | XCR1 | 1422294_at | 9440 | 0.062 | -0.1960 | No | ||
| 40 | PARP4 | 1441026_at | 9481 | 0.059 | -0.1970 | No | ||
| 41 | TNFRSF1A | 1417291_at | 9678 | 0.046 | -0.2054 | No | ||
| 42 | NFATC4 | 1423380_s_at 1432821_at 1454369_a_at | 9710 | 0.045 | -0.2062 | No | ||
| 43 | C3AR1 | 1419482_at 1419483_at 1442082_at | 9998 | 0.026 | -0.2190 | No | ||
| 44 | CCR1 | 1419609_at 1419610_at | 10082 | 0.020 | -0.2225 | No | ||
| 45 | NOX4 | 1419161_a_at 1451827_a_at | 10386 | 0.001 | -0.2364 | No | ||
| 46 | ELF3 | 1416916_at | 10569 | -0.010 | -0.2446 | No | ||
| 47 | IL1RAP | 1421843_at 1439697_at 1442614_at 1449585_at | 10783 | -0.023 | -0.2541 | No | ||
| 48 | TPST1 | 1421733_a_at | 11216 | -0.051 | -0.2732 | No | ||
| 49 | IL1A | 1421473_at | 11680 | -0.080 | -0.2933 | No | ||
| 50 | SCG2 | 1450708_at | 12590 | -0.135 | -0.3331 | No | ||
| 51 | ORM2 | 1420438_at | 13124 | -0.170 | -0.3553 | No | ||
| 52 | RIPK2 | 1421236_at 1450173_at | 13249 | -0.177 | -0.3585 | No | ||
| 53 | HDAC7A | 1420812_at 1420813_at | 13271 | -0.179 | -0.3571 | No | ||
| 54 | CXCL2 | 1449984_at | 13371 | -0.186 | -0.3591 | No | ||
| 55 | IL20 | 1421608_at | 13888 | -0.221 | -0.3798 | No | ||
| 56 | CXCL11 | 1419697_at 1419698_at | 13915 | -0.223 | -0.3779 | No | ||
| 57 | SCYE1 | 1416486_at | 14135 | -0.237 | -0.3848 | No | ||
| 58 | GHRL | 1448980_at | 14238 | -0.244 | -0.3861 | No | ||
| 59 | TACR1 | 1422282_at | 14349 | -0.252 | -0.3878 | No | ||
| 60 | HDAC9 | 1421306_a_at 1434572_at | 14480 | -0.261 | -0.3902 | No | ||
| 61 | IFNA2 | 1423028_at | 15019 | -0.301 | -0.4108 | No | ||
| 62 | CCL20 | 1422029_at | 15181 | -0.313 | -0.4139 | No | ||
| 63 | ORM1 | 1451054_at | 15213 | -0.316 | -0.4110 | No | ||
| 64 | LTB4R | 1420407_at | 15296 | -0.322 | -0.4104 | No | ||
| 65 | AHSG | 1455093_a_at | 15478 | -0.336 | -0.4142 | No | ||
| 66 | PTAFR | 1427871_at 1427872_at | 15747 | -0.355 | -0.4217 | No | ||
| 67 | NMI | 1425719_a_at | 15757 | -0.356 | -0.4173 | No | ||
| 68 | MBL2 | 1418787_at | 16381 | -0.409 | -0.4403 | No | ||
| 69 | C2 | 1416051_at 1441912_x_at 1457664_x_at | 16605 | -0.430 | -0.4447 | Yes | ||
| 70 | F11R | 1424595_at 1436374_x_at | 16709 | -0.438 | -0.4435 | Yes | ||
| 71 | ALOX15 | 1420338_at | 16776 | -0.445 | -0.4405 | Yes | ||
| 72 | ABCF1 | 1420157_s_at 1420158_s_at 1426921_at 1427444_at 1452236_at 1452429_s_at | 16884 | -0.457 | -0.4392 | Yes | ||
| 73 | LBP | 1448550_at | 16998 | -0.469 | -0.4380 | Yes | ||
| 74 | PTX3 | 1418666_at 1456903_at | 17192 | -0.488 | -0.4403 | Yes | ||
| 75 | BLNK | 1451780_at | 17243 | -0.494 | -0.4359 | Yes | ||
| 76 | CD40LG | 1422283_at | 17543 | -0.526 | -0.4425 | Yes | ||
| 77 | CXCR4 | 1448710_at | 17598 | -0.531 | -0.4377 | Yes | ||
| 78 | ALOX5AP | 1452016_at 1459234_at | 17637 | -0.537 | -0.4322 | Yes | ||
| 79 | IL18RAP | 1421291_at 1456545_at | 17842 | -0.562 | -0.4340 | Yes | ||
| 80 | CCR3 | 1422957_at | 18034 | -0.588 | -0.4348 | Yes | ||
| 81 | CXCL9 | 1418652_at 1456907_at | 18124 | -0.601 | -0.4307 | Yes | ||
| 82 | PRDX5 | 1416381_a_at | 18230 | -0.614 | -0.4272 | Yes | ||
| 83 | NFKB1 | 1427705_a_at 1442949_at | 18289 | -0.622 | -0.4214 | Yes | ||
| 84 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 18330 | -0.627 | -0.4148 | Yes | ||
| 85 | CCL11 | 1417789_at | 18373 | -0.633 | -0.4081 | Yes | ||
| 86 | SELE | 1421712_at | 18378 | -0.634 | -0.3998 | Yes | ||
| 87 | IL13 | 1420802_at | 18588 | -0.666 | -0.4003 | Yes | ||
| 88 | RELA | 1419536_a_at | 18597 | -0.667 | -0.3917 | Yes | ||
| 89 | CXCL10 | 1418930_at | 18611 | -0.669 | -0.3832 | Yes | ||
| 90 | NFRKB | 1434753_at 1445551_at | 18921 | -0.717 | -0.3877 | Yes | ||
| 91 | CRP | 1421946_at | 19231 | -0.772 | -0.3914 | Yes | ||
| 92 | CHST2 | 1422758_at 1460070_at | 19759 | -0.875 | -0.4037 | Yes | ||
| 93 | AIF1 | 1418204_s_at | 19980 | -0.928 | -0.4012 | Yes | ||
| 94 | AOAH | 1450764_at 1460156_at | 20293 | -1.003 | -0.4019 | Yes | ||
| 95 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 20376 | -1.019 | -0.3919 | Yes | ||
| 96 | NFE2L1 | 1416331_a_at | 20700 | -1.109 | -0.3917 | Yes | ||
| 97 | SIGIRR | 1449163_at | 20982 | -1.245 | -0.3877 | Yes | ||
| 98 | IL10RB | 1419455_at | 21001 | -1.256 | -0.3716 | Yes | ||
| 99 | HDAC5 | 1415743_at | 21049 | -1.281 | -0.3564 | Yes | ||
| 100 | GPR68 | 1455000_at | 21315 | -1.468 | -0.3486 | Yes | ||
| 101 | IRAK2 | 1436507_at | 21594 | -1.807 | -0.3369 | Yes | ||
| 102 | S100A8 | 1419394_s_at | 21598 | -1.810 | -0.3126 | Yes | ||
| 103 | MGLL | 1426785_s_at 1431331_at 1450391_a_at 1453836_a_at | 21652 | -1.930 | -0.2889 | Yes | ||
| 104 | CD40 | 1439221_s_at 1449473_s_at 1460415_a_at | 21666 | -1.970 | -0.2628 | Yes | ||
| 105 | S100A9 | 1448756_at | 21769 | -2.327 | -0.2360 | Yes | ||
| 106 | PLA2G2D | 1418987_at 1454157_a_at | 21806 | -2.622 | -0.2022 | Yes | ||
| 107 | CCR4 | 1421655_a_at | 21867 | -3.180 | -0.1619 | Yes | ||
| 108 | CCL22 | 1417925_at | 21871 | -3.238 | -0.1182 | Yes | ||
| 109 | FOS | 1423100_at | 21901 | -3.644 | -0.0702 | Yes | ||
| 110 | CCR7 | 1423466_at | 21924 | -5.296 | 0.0005 | Yes |

