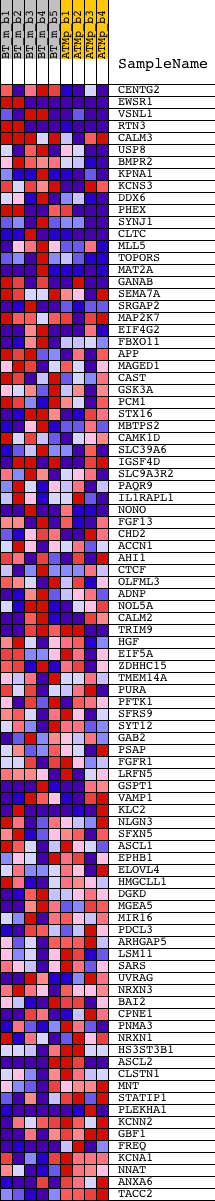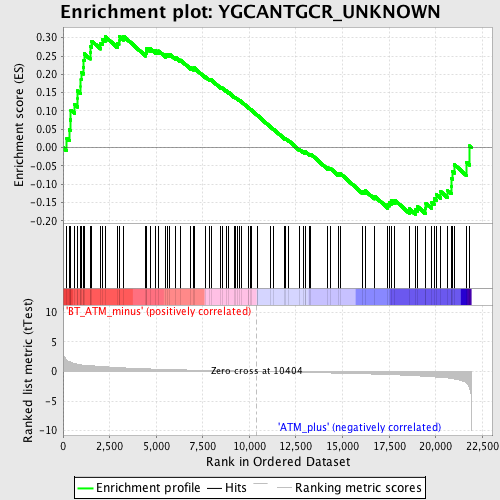
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | YGCANTGCR_UNKNOWN |
| Enrichment Score (ES) | 0.30432194 |
| Normalized Enrichment Score (NES) | 1.2357327 |
| Nominal p-value | 0.1025 |
| FDR q-value | 0.5465454 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CENTG2 | 1435432_at 1435433_at 1437394_at 1442632_at 1457978_at 1459053_at 1459389_at | 200 | 1.897 | 0.0252 | Yes | ||
| 2 | EWSR1 | 1417238_at 1425919_at 1436884_x_at 1441074_at 1456461_at | 342 | 1.629 | 0.0483 | Yes | ||
| 3 | VSNL1 | 1420955_at 1450055_at | 380 | 1.602 | 0.0756 | Yes | ||
| 4 | RTN3 | 1418101_a_at 1425539_a_at 1442365_at 1443220_at | 418 | 1.561 | 0.1022 | Yes | ||
| 5 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 608 | 1.338 | 0.1178 | Yes | ||
| 6 | USP8 | 1422883_at 1443420_at 1446807_at 1447386_at | 769 | 1.229 | 0.1328 | Yes | ||
| 7 | BMPR2 | 1419616_at 1434310_at 1441652_at | 774 | 1.225 | 0.1548 | Yes | ||
| 8 | KPNA1 | 1419548_at 1449503_at 1449504_at 1460260_s_at | 957 | 1.116 | 0.1667 | Yes | ||
| 9 | KCNS3 | 1445519_at 1457799_at | 959 | 1.116 | 0.1869 | Yes | ||
| 10 | DDX6 | 1424598_at 1457618_at | 1000 | 1.093 | 0.2049 | Yes | ||
| 11 | PHEX | 1421979_at 1450445_at | 1092 | 1.062 | 0.2199 | Yes | ||
| 12 | SYNJ1 | 1436333_a_at 1436334_at 1439672_at 1439997_at 1454961_at | 1114 | 1.056 | 0.2381 | Yes | ||
| 13 | CLTC | 1419407_at 1439094_at 1440457_at 1454626_at | 1139 | 1.047 | 0.2560 | Yes | ||
| 14 | MLL5 | 1427236_a_at 1432601_at 1434704_at 1439107_a_at 1439108_at | 1451 | 0.979 | 0.2595 | Yes | ||
| 15 | TOPORS | 1417754_at 1417755_at 1448838_at | 1463 | 0.977 | 0.2767 | Yes | ||
| 16 | MAT2A | 1423667_at 1433576_at 1438386_x_at 1438630_x_at 1438976_x_at 1439386_x_at 1456702_x_at | 1534 | 0.961 | 0.2909 | Yes | ||
| 17 | GANAB | 1415787_at 1437812_x_at | 2012 | 0.852 | 0.2845 | Yes | ||
| 18 | SEMA7A | 1422040_at 1459903_at | 2104 | 0.832 | 0.2955 | Yes | ||
| 19 | SRGAP2 | 1427967_at 1429884_at 1434406_at 1434407_at | 2256 | 0.797 | 0.3030 | Yes | ||
| 20 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 2920 | 0.662 | 0.2846 | Yes | ||
| 21 | EIF4G2 | 1415863_at 1452758_s_at | 3002 | 0.646 | 0.2926 | Yes | ||
| 22 | FBXO11 | 1434108_at | 3007 | 0.644 | 0.3041 | Yes | ||
| 23 | APP | 1420621_a_at 1427442_a_at 1438373_at 1438374_x_at 1444705_at 1447347_at 1458525_at | 3242 | 0.603 | 0.3043 | Yes | ||
| 24 | MAGED1 | 1450062_a_at | 4433 | 0.447 | 0.2579 | No | ||
| 25 | CAST | 1426098_a_at 1435972_at 1451413_at | 4479 | 0.442 | 0.2639 | No | ||
| 26 | GSK3A | 1435638_at | 4494 | 0.440 | 0.2712 | No | ||
| 27 | PCM1 | 1418524_at 1418525_at 1431287_at 1436908_at 1438528_at 1446140_at 1449120_a_at | 4680 | 0.422 | 0.2704 | No | ||
| 28 | STX16 | 1429237_at | 4964 | 0.394 | 0.2646 | No | ||
| 29 | MBTPS2 | 1436883_at 1437495_at | 5132 | 0.377 | 0.2638 | No | ||
| 30 | CAMK1D | 1426389_at 1438643_at 1452050_at | 5506 | 0.344 | 0.2530 | No | ||
| 31 | SLC39A6 | 1424674_at 1424675_at 1441949_x_at 1447893_x_at | 5615 | 0.335 | 0.2541 | No | ||
| 32 | IGSF4D | 1435145_at 1435146_s_at 1435147_x_at 1436743_at 1440412_at 1444664_at 1459761_x_at | 5725 | 0.324 | 0.2550 | No | ||
| 33 | SLC9A3R2 | 1428954_at 1428955_x_at 1431208_a_at 1439368_a_at 1439369_x_at 1452976_a_at | 6032 | 0.297 | 0.2464 | No | ||
| 34 | PAQR9 | 1455025_at | 6301 | 0.277 | 0.2391 | No | ||
| 35 | IL1RAPL1 | 1445821_at 1458964_at | 6865 | 0.233 | 0.2175 | No | ||
| 36 | NONO | 1415820_x_at 1431239_at 1448103_s_at | 6998 | 0.223 | 0.2155 | No | ||
| 37 | FGF13 | 1418497_at 1418498_at | 7036 | 0.220 | 0.2178 | No | ||
| 38 | CHD2 | 1444246_at 1445843_at | 7648 | 0.178 | 0.1931 | No | ||
| 39 | ACCN1 | 1417994_a_at | 7850 | 0.164 | 0.1868 | No | ||
| 40 | AHI1 | 1439083_at 1455177_at | 7953 | 0.158 | 0.1850 | No | ||
| 41 | CTCF | 1418330_at 1449042_at | 8463 | 0.123 | 0.1640 | No | ||
| 42 | OLFML3 | 1448475_at | 8537 | 0.118 | 0.1627 | No | ||
| 43 | ADNP | 1423069_at | 8747 | 0.104 | 0.1551 | No | ||
| 44 | NOL5A | 1426533_at 1455035_s_at | 8907 | 0.095 | 0.1495 | No | ||
| 45 | CALM2 | 1422414_a_at 1423807_a_at | 9204 | 0.077 | 0.1373 | No | ||
| 46 | TRIM9 | 1434595_at 1443989_at | 9237 | 0.075 | 0.1372 | No | ||
| 47 | HGF | 1425379_at 1442884_at 1451866_a_at | 9381 | 0.065 | 0.1319 | No | ||
| 48 | EIF5A | 1437859_x_at 1451470_s_at | 9451 | 0.061 | 0.1298 | No | ||
| 49 | ZDHHC15 | 1442274_at | 9562 | 0.054 | 0.1258 | No | ||
| 50 | TMEM14A | 1428447_at | 9934 | 0.030 | 0.1093 | No | ||
| 51 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 9977 | 0.027 | 0.1079 | No | ||
| 52 | PFTK1 | 1419249_at 1419250_a_at 1441692_at 1445984_at 1453956_a_at | 10079 | 0.020 | 0.1037 | No | ||
| 53 | SFRS9 | 1417727_at | 10101 | 0.019 | 0.1030 | No | ||
| 54 | SYT12 | 1422878_at | 10413 | -0.000 | 0.0888 | No | ||
| 55 | GAB2 | 1419829_a_at 1420785_at 1439786_at 1446860_at | 10443 | -0.002 | 0.0875 | No | ||
| 56 | PSAP | 1415687_a_at 1421813_a_at | 11152 | -0.047 | 0.0559 | No | ||
| 57 | FGFR1 | 1424050_s_at 1425911_a_at 1436551_at | 11314 | -0.057 | 0.0496 | No | ||
| 58 | LRFN5 | 1441502_at 1454687_at | 11910 | -0.094 | 0.0241 | No | ||
| 59 | GSPT1 | 1426736_at 1426737_at 1435000_at 1446550_at 1452168_x_at 1455173_at | 11964 | -0.097 | 0.0234 | No | ||
| 60 | VAMP1 | 1421862_a_at 1421863_at | 12093 | -0.104 | 0.0194 | No | ||
| 61 | KLC2 | 1418214_at 1431487_at | 12678 | -0.140 | -0.0048 | No | ||
| 62 | NLGN3 | 1456384_at | 12897 | -0.155 | -0.0119 | No | ||
| 63 | SFXN5 | 1436618_at 1440428_at | 13005 | -0.161 | -0.0139 | No | ||
| 64 | ASCL1 | 1437086_at 1450164_at | 13009 | -0.161 | -0.0111 | No | ||
| 65 | EPHB1 | 1455188_at | 13254 | -0.178 | -0.0191 | No | ||
| 66 | ELOVL4 | 1424306_at 1451308_at | 13298 | -0.181 | -0.0178 | No | ||
| 67 | HMGCLL1 | 1434913_at | 14218 | -0.242 | -0.0555 | No | ||
| 68 | DGKD | 1433564_at 1439873_at 1442254_at 1454655_at | 14339 | -0.251 | -0.0564 | No | ||
| 69 | MGEA5 | 1422900_at 1422901_at 1422902_s_at 1433654_at 1439081_at | 14763 | -0.283 | -0.0706 | No | ||
| 70 | MIR16 | 1418444_a_at | 14871 | -0.290 | -0.0703 | No | ||
| 71 | PDCL3 | 1424002_at | 16096 | -0.385 | -0.1193 | No | ||
| 72 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 16227 | -0.396 | -0.1181 | No | ||
| 73 | LSM11 | 1453755_at | 16718 | -0.439 | -0.1326 | No | ||
| 74 | SARS | 1426257_a_at 1452000_s_at | 17440 | -0.515 | -0.1563 | No | ||
| 75 | UVRAG | 1454706_at | 17516 | -0.523 | -0.1502 | No | ||
| 76 | NRXN3 | 1419825_at 1431153_at 1432931_at 1433788_at 1438193_at 1439629_at 1442423_at 1444700_at 1456137_at 1460101_at | 17615 | -0.534 | -0.1450 | No | ||
| 77 | BAI2 | 1435558_at | 17814 | -0.558 | -0.1440 | No | ||
| 78 | CPNE1 | 1433439_at | 18590 | -0.666 | -0.1674 | No | ||
| 79 | PNMA3 | 1456829_at | 18928 | -0.718 | -0.1698 | No | ||
| 80 | NRXN1 | 1428240_at 1433413_at 1439358_a_at 1439359_x_at 1442501_at 1447216_at 1454691_at 1460540_at | 19034 | -0.735 | -0.1613 | No | ||
| 81 | HS3ST3B1 | 1421331_at 1421332_at 1433977_at 1442816_at 1444055_at | 19448 | -0.811 | -0.1655 | No | ||
| 82 | ASCL2 | 1422396_s_at 1432018_at 1460514_s_at | 19482 | -0.817 | -0.1522 | No | ||
| 83 | CLSTN1 | 1421860_at 1421861_at 1445085_at | 19801 | -0.885 | -0.1507 | No | ||
| 84 | MNT | 1418192_at | 19924 | -0.914 | -0.1397 | No | ||
| 85 | STATIP1 | 1415774_at 1438179_s_at | 20041 | -0.943 | -0.1280 | No | ||
| 86 | PLEKHA1 | 1417965_at 1443111_at | 20283 | -1.001 | -0.1208 | No | ||
| 87 | KCNN2 | 1445676_at 1448927_at | 20634 | -1.088 | -0.1171 | No | ||
| 88 | GBF1 | 1415709_s_at 1415757_at 1432677_at 1438207_at | 20847 | -1.177 | -0.1055 | No | ||
| 89 | FREQ | 1434887_at 1450146_at 1460293_at | 20856 | -1.182 | -0.0845 | No | ||
| 90 | KCNA1 | 1417416_at 1437230_at 1442413_at 1455785_at | 20914 | -1.213 | -0.0651 | No | ||
| 91 | NNAT | 1423506_a_at | 21018 | -1.263 | -0.0469 | No | ||
| 92 | ANXA6 | 1415818_at 1429246_a_at 1429247_at 1442288_at | 21677 | -2.002 | -0.0407 | No | ||
| 93 | TACC2 | 1425745_a_at 1447891_at | 21836 | -2.894 | 0.0045 | No |