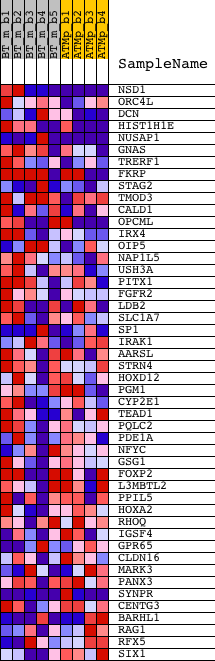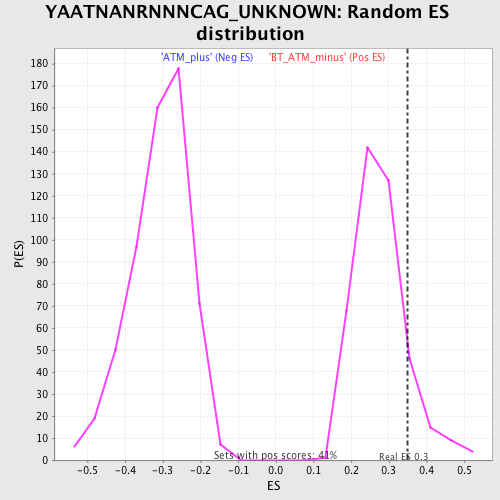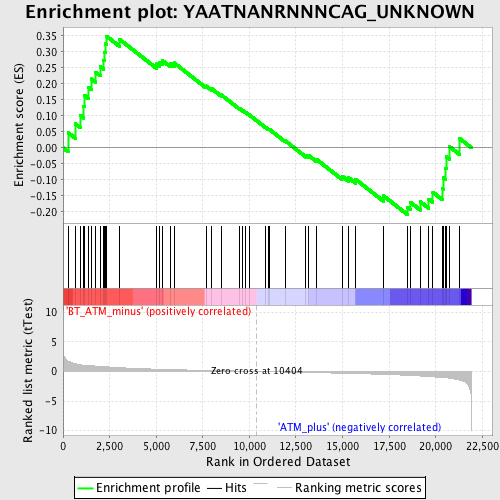
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | YAATNANRNNNCAG_UNKNOWN |
| Enrichment Score (ES) | 0.3490536 |
| Normalized Enrichment Score (NES) | 1.2578144 |
| Nominal p-value | 0.11407767 |
| FDR q-value | 0.534077 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NSD1 | 1420881_at 1435088_at 1440972_at 1456916_at | 266 | 1.739 | 0.0478 | Yes | ||
| 2 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 648 | 1.303 | 0.0753 | Yes | ||
| 3 | DCN | 1441506_at 1449368_at | 936 | 1.127 | 0.1010 | Yes | ||
| 4 | HIST1H1E | 1422398_at 1440330_at 1443209_at 1445565_at | 1110 | 1.056 | 0.1294 | Yes | ||
| 5 | NUSAP1 | 1416309_at 1416310_at 1442830_at | 1137 | 1.048 | 0.1644 | Yes | ||
| 6 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 1347 | 1.000 | 0.1892 | Yes | ||
| 7 | TRERF1 | 1444654_at | 1507 | 0.967 | 0.2153 | Yes | ||
| 8 | FKRP | 1436812_at 1437536_at 1438414_at | 1734 | 0.918 | 0.2366 | Yes | ||
| 9 | STAG2 | 1421849_at 1450396_at | 1991 | 0.857 | 0.2544 | Yes | ||
| 10 | TMOD3 | 1423088_at 1423089_at 1438556_a_at 1439626_at 1455708_at 1456913_at | 2180 | 0.812 | 0.2738 | Yes | ||
| 11 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 2233 | 0.801 | 0.2990 | Yes | ||
| 12 | OPCML | 1435612_at 1442752_at 1457446_at 1458228_at | 2272 | 0.794 | 0.3246 | Yes | ||
| 13 | IRX4 | 1419539_at | 2327 | 0.781 | 0.3491 | Yes | ||
| 14 | OIP5 | 1430617_at | 3024 | 0.642 | 0.3394 | No | ||
| 15 | NAP1L5 | 1417411_at | 5008 | 0.390 | 0.2622 | No | ||
| 16 | USH3A | 1438347_at | 5172 | 0.374 | 0.2676 | No | ||
| 17 | PITX1 | 1419514_at 1449488_at | 5332 | 0.360 | 0.2727 | No | ||
| 18 | FGFR2 | 1420847_a_at 1433489_s_at 1443996_at | 5761 | 0.321 | 0.2642 | No | ||
| 19 | LDB2 | 1421101_a_at 1439557_s_at 1445126_at 1456786_at | 5967 | 0.303 | 0.2653 | No | ||
| 20 | SLC1A7 | 1456971_at | 7676 | 0.175 | 0.1933 | No | ||
| 21 | SP1 | 1418180_at 1448994_at 1454852_at | 7990 | 0.155 | 0.1843 | No | ||
| 22 | IRAK1 | 1448668_a_at 1460649_at | 8492 | 0.121 | 0.1656 | No | ||
| 23 | AARSL | 1436017_at | 9496 | 0.059 | 0.1218 | No | ||
| 24 | STRN4 | 1426209_at | 9650 | 0.048 | 0.1165 | No | ||
| 25 | HOXD12 | 1450553_at | 9810 | 0.039 | 0.1106 | No | ||
| 26 | PGM1 | 1446824_at 1453283_at | 9991 | 0.027 | 0.1033 | No | ||
| 27 | CYP2E1 | 1415994_at | 10891 | -0.031 | 0.0633 | No | ||
| 28 | TEAD1 | 1423434_at 1447911_at 1456717_at | 11016 | -0.039 | 0.0590 | No | ||
| 29 | PQLC2 | 1425632_a_at | 11071 | -0.042 | 0.0579 | No | ||
| 30 | PDE1A | 1431625_at 1449298_a_at | 11929 | -0.095 | 0.0220 | No | ||
| 31 | NFYC | 1448963_at | 13041 | -0.163 | -0.0231 | No | ||
| 32 | GSG1 | 1417550_a_at | 13182 | -0.173 | -0.0235 | No | ||
| 33 | FOXP2 | 1422014_at 1438231_at 1438232_at 1440108_at 1441365_at 1458191_at | 13585 | -0.200 | -0.0350 | No | ||
| 34 | L3MBTL2 | 1433777_at | 14980 | -0.299 | -0.0884 | No | ||
| 35 | PPIL5 | 1427506_at 1436922_at 1452458_s_at | 15331 | -0.324 | -0.0933 | No | ||
| 36 | HOXA2 | 1419602_at | 15712 | -0.353 | -0.0985 | No | ||
| 37 | RHOQ | 1427918_a_at 1440925_at | 17204 | -0.489 | -0.1497 | No | ||
| 38 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 18481 | -0.649 | -0.1857 | No | ||
| 39 | GPR65 | 1449175_at | 18652 | -0.675 | -0.1702 | No | ||
| 40 | CLDN16 | 1420434_at | 19182 | -0.761 | -0.1682 | No | ||
| 41 | MARK3 | 1418316_a_at | 19640 | -0.850 | -0.1598 | No | ||
| 42 | PANX3 | 1432860_at 1454387_at 1456073_s_at | 19859 | -0.898 | -0.1388 | No | ||
| 43 | SYNPR | 1423640_at 1444331_at | 20359 | -1.015 | -0.1266 | No | ||
| 44 | CENTG3 | 1423910_at 1434032_at | 20408 | -1.027 | -0.0934 | No | ||
| 45 | BARHL1 | 1418811_at 1418812_a_at | 20544 | -1.061 | -0.0631 | No | ||
| 46 | RAG1 | 1440574_at 1450680_at | 20591 | -1.074 | -0.0282 | No | ||
| 47 | RFX5 | 1423103_at | 20763 | -1.139 | 0.0033 | No | ||
| 48 | SIX1 | 1427277_at | 21295 | -1.456 | 0.0292 | No |