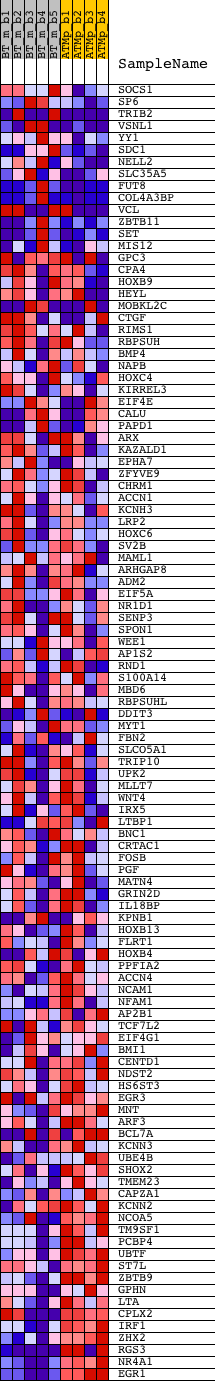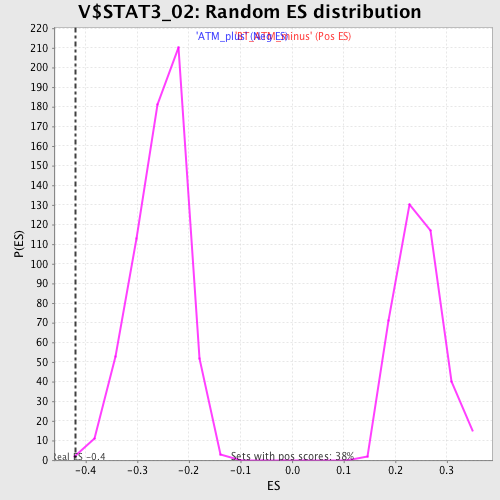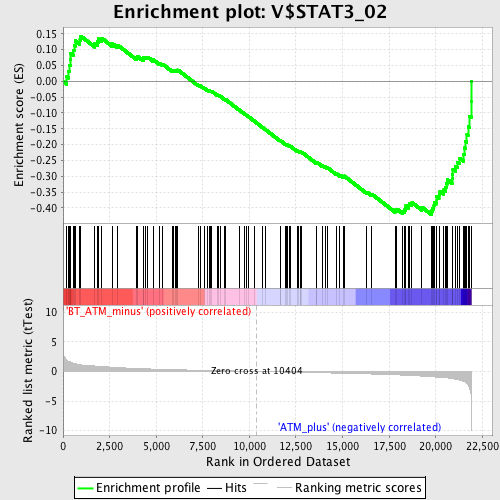
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$STAT3_02 |
| Enrichment Score (ES) | -0.4206122 |
| Normalized Enrichment Score (NES) | -1.6474326 |
| Nominal p-value | 0.0016 |
| FDR q-value | 0.044613406 |
| FWER p-Value | 0.465 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SOCS1 | 1440047_at 1450446_a_at | 198 | 1.904 | 0.0151 | No | ||
| 2 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 305 | 1.666 | 0.0313 | No | ||
| 3 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 346 | 1.628 | 0.0502 | No | ||
| 4 | VSNL1 | 1420955_at 1450055_at | 380 | 1.602 | 0.0690 | No | ||
| 5 | YY1 | 1422569_at 1422570_at 1435824_at 1457834_at | 413 | 1.567 | 0.0874 | No | ||
| 6 | SDC1 | 1415943_at 1415944_at 1437279_x_at 1448158_at | 569 | 1.365 | 0.0976 | No | ||
| 7 | NELL2 | 1423560_at 1423561_at | 595 | 1.348 | 0.1135 | No | ||
| 8 | SLC35A5 | 1419970_at 1419971_s_at 1419972_at 1423457_at 1423458_at | 644 | 1.308 | 0.1279 | No | ||
| 9 | FUT8 | 1441015_at 1442483_at 1443656_at 1456367_at 1460319_at | 905 | 1.142 | 0.1305 | No | ||
| 10 | COL4A3BP | 1420383_a_at 1420384_at 1449847_a_at 1452867_at | 950 | 1.121 | 0.1427 | No | ||
| 11 | VCL | 1416156_at 1416157_at 1445256_at | 1706 | 0.924 | 0.1198 | No | ||
| 12 | ZBTB11 | 1454826_at | 1835 | 0.894 | 0.1253 | No | ||
| 13 | SET | 1426853_at 1426854_a_at | 1892 | 0.881 | 0.1339 | No | ||
| 14 | MIS12 | 1424717_at | 2083 | 0.839 | 0.1358 | No | ||
| 15 | GPC3 | 1443240_at 1446525_at 1450990_at | 2664 | 0.706 | 0.1182 | No | ||
| 16 | CPA4 | 1453770_at | 2930 | 0.659 | 0.1144 | No | ||
| 17 | HOXB9 | 1452317_at | 3932 | 0.507 | 0.0750 | No | ||
| 18 | HEYL | 1419302_at 1419303_at 1438886_at | 3985 | 0.501 | 0.0790 | No | ||
| 19 | MOBKL2C | 1440919_at 1459919_a_at | 4303 | 0.461 | 0.0703 | No | ||
| 20 | CTGF | 1416953_at | 4313 | 0.461 | 0.0757 | No | ||
| 21 | RIMS1 | 1435667_at 1438305_at 1444393_at | 4420 | 0.449 | 0.0766 | No | ||
| 22 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 4547 | 0.435 | 0.0763 | No | ||
| 23 | BMP4 | 1422912_at | 4840 | 0.406 | 0.0681 | No | ||
| 24 | NAPB | 1423172_at 1423173_at 1427470_s_at 1450888_at 1452444_at | 5201 | 0.371 | 0.0563 | No | ||
| 25 | HOXC4 | 1422870_at | 5361 | 0.356 | 0.0535 | No | ||
| 26 | KIRREL3 | 1431402_at 1441357_at 1452728_at | 5855 | 0.311 | 0.0349 | No | ||
| 27 | EIF4E | 1450909_at 1457489_at 1459371_at | 5942 | 0.305 | 0.0348 | No | ||
| 28 | CALU | 1415870_at 1441477_at | 6061 | 0.295 | 0.0332 | No | ||
| 29 | PAPD1 | 1452749_at | 6110 | 0.292 | 0.0347 | No | ||
| 30 | ARX | 1420926_at 1441175_at 1450042_at | 6167 | 0.287 | 0.0357 | No | ||
| 31 | KAZALD1 | 1436528_at | 7263 | 0.204 | -0.0118 | No | ||
| 32 | EPHA7 | 1427528_a_at 1443485_at 1445635_at 1451991_at 1452380_at | 7356 | 0.198 | -0.0135 | No | ||
| 33 | ZFYVE9 | 1440348_at | 7577 | 0.183 | -0.0213 | No | ||
| 34 | CHRM1 | 1439611_at 1450833_at | 7765 | 0.170 | -0.0277 | No | ||
| 35 | ACCN1 | 1417994_a_at | 7850 | 0.164 | -0.0295 | No | ||
| 36 | KCNH3 | 1421619_at 1431690_at 1445805_x_at 1459107_at | 7934 | 0.159 | -0.0313 | No | ||
| 37 | LRP2 | 1427133_s_at 1427621_at 1445901_at 1452320_at | 7976 | 0.156 | -0.0312 | No | ||
| 38 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 8275 | 0.134 | -0.0431 | No | ||
| 39 | SV2B | 1434800_at 1435687_at | 8326 | 0.131 | -0.0437 | No | ||
| 40 | MAML1 | 1426769_s_at 1441697_at 1445045_at 1452188_at | 8446 | 0.123 | -0.0476 | No | ||
| 41 | ARHGAP8 | 1451463_at | 8688 | 0.108 | -0.0573 | No | ||
| 42 | ADM2 | 1442856_at | 8745 | 0.105 | -0.0585 | No | ||
| 43 | EIF5A | 1437859_x_at 1451470_s_at | 9451 | 0.061 | -0.0901 | No | ||
| 44 | NR1D1 | 1421802_at 1426464_at | 9713 | 0.045 | -0.1014 | No | ||
| 45 | SENP3 | 1448866_at | 9854 | 0.035 | -0.1074 | No | ||
| 46 | SPON1 | 1424415_s_at 1441226_at 1451342_at | 9931 | 0.030 | -0.1105 | No | ||
| 47 | WEE1 | 1416773_at 1416774_at | 10282 | 0.008 | -0.1264 | No | ||
| 48 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 10713 | -0.018 | -0.1459 | No | ||
| 49 | RND1 | 1455197_at | 10855 | -0.029 | -0.1520 | No | ||
| 50 | S100A14 | 1449166_at | 11646 | -0.077 | -0.1872 | No | ||
| 51 | MBD6 | 1418375_at | 11664 | -0.079 | -0.1870 | No | ||
| 52 | RBPSUHL | 1421956_at 1450433_at | 11955 | -0.096 | -0.1991 | No | ||
| 53 | DDIT3 | 1417516_at 1443897_at | 12017 | -0.099 | -0.2006 | No | ||
| 54 | MYT1 | 1422773_at 1439365_at | 12048 | -0.102 | -0.2007 | No | ||
| 55 | FBN2 | 1454830_at | 12139 | -0.107 | -0.2035 | No | ||
| 56 | SLCO5A1 | 1439588_at 1440874_at | 12225 | -0.112 | -0.2059 | No | ||
| 57 | TRIP10 | 1418092_s_at | 12569 | -0.134 | -0.2199 | No | ||
| 58 | UPK2 | 1421781_at | 12636 | -0.138 | -0.2212 | No | ||
| 59 | MLLT7 | 1422149_at | 12721 | -0.143 | -0.2233 | No | ||
| 60 | WNT4 | 1450782_at | 12722 | -0.144 | -0.2214 | No | ||
| 61 | IRX5 | 1421072_at | 12809 | -0.149 | -0.2235 | No | ||
| 62 | LTBP1 | 1419786_at 1440678_at 1446232_at 1447547_at 1448870_at 1457852_at 1458739_at | 13621 | -0.202 | -0.2581 | No | ||
| 63 | BNC1 | 1424890_at | 13627 | -0.203 | -0.2557 | No | ||
| 64 | CRTAC1 | 1426606_at 1452936_at 1457474_at 1459858_x_at | 13930 | -0.224 | -0.2667 | No | ||
| 65 | FOSB | 1422134_at | 14066 | -0.232 | -0.2700 | No | ||
| 66 | PGF | 1418471_at | 14171 | -0.239 | -0.2717 | No | ||
| 67 | MATN4 | 1418464_at | 14666 | -0.276 | -0.2908 | No | ||
| 68 | GRIN2D | 1421393_at | 14868 | -0.290 | -0.2964 | No | ||
| 69 | IL18BP | 1450424_a_at | 15051 | -0.304 | -0.3008 | No | ||
| 70 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 15097 | -0.307 | -0.2990 | No | ||
| 71 | HOXB13 | 1419576_at | 16314 | -0.403 | -0.3496 | No | ||
| 72 | FLRT1 | 1443143_at | 16580 | -0.427 | -0.3563 | No | ||
| 73 | HOXB4 | 1451761_at 1460379_at | 17824 | -0.559 | -0.4062 | No | ||
| 74 | PPFIA2 | 1445766_at 1446218_at 1456856_at | 17925 | -0.574 | -0.4035 | No | ||
| 75 | ACCN4 | 1437091_at | 18241 | -0.615 | -0.4101 | Yes | ||
| 76 | NCAM1 | 1421966_at 1425126_at 1426864_a_at 1426865_a_at 1439556_at 1440067_at 1441995_at 1442680_at 1443018_at 1450437_a_at 1450438_at 1454140_at | 18333 | -0.627 | -0.4063 | Yes | ||
| 77 | NFAM1 | 1425714_a_at 1428790_at | 18382 | -0.634 | -0.4005 | Yes | ||
| 78 | AP2B1 | 1427077_a_at 1445142_at 1446566_at 1452292_at 1458971_at | 18410 | -0.638 | -0.3936 | Yes | ||
| 79 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 18563 | -0.662 | -0.3922 | Yes | ||
| 80 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 18593 | -0.666 | -0.3851 | Yes | ||
| 81 | BMI1 | 1417493_at 1448733_at | 18733 | -0.689 | -0.3827 | Yes | ||
| 82 | CENTD1 | 1436895_at 1446649_at 1452291_at 1456337_at 1458173_at | 19268 | -0.779 | -0.3973 | Yes | ||
| 83 | NDST2 | 1417931_at | 19778 | -0.881 | -0.4094 | Yes | ||
| 84 | HS6ST3 | 1421599_at | 19850 | -0.897 | -0.4013 | Yes | ||
| 85 | EGR3 | 1421486_at 1436329_at | 19914 | -0.912 | -0.3926 | Yes | ||
| 86 | MNT | 1418192_at | 19924 | -0.914 | -0.3815 | Yes | ||
| 87 | ARF3 | 1421789_s_at 1423973_a_at 1434787_at 1437331_a_at 1456007_at | 20058 | -0.947 | -0.3755 | Yes | ||
| 88 | BCL7A | 1428207_at 1428208_at 1451855_at 1459416_at | 20060 | -0.949 | -0.3636 | Yes | ||
| 89 | KCNN3 | 1421632_at 1459308_at | 20187 | -0.977 | -0.3569 | Yes | ||
| 90 | UBE4B | 1432499_a_at 1444465_at 1448410_at | 20236 | -0.993 | -0.3465 | Yes | ||
| 91 | SHOX2 | 1420559_a_at 1438042_at | 20450 | -1.036 | -0.3432 | Yes | ||
| 92 | TMEM23 | 1426575_at 1426576_at 1436499_at 1442079_at 1445103_at 1457968_at | 20548 | -1.062 | -0.3341 | Yes | ||
| 93 | CAPZA1 | 1439455_x_at 1440380_at | 20599 | -1.076 | -0.3228 | Yes | ||
| 94 | KCNN2 | 1445676_at 1448927_at | 20634 | -1.088 | -0.3105 | Yes | ||
| 95 | NCOA5 | 1424055_at 1456997_at | 20887 | -1.195 | -0.3069 | Yes | ||
| 96 | TM9SF1 | 1418198_a_at 1442908_at | 20917 | -1.215 | -0.2928 | Yes | ||
| 97 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20933 | -1.222 | -0.2780 | Yes | ||
| 98 | UBTF | 1453097_a_at 1455490_at 1460304_a_at | 21076 | -1.300 | -0.2680 | Yes | ||
| 99 | ST7L | 1427916_at 1456363_at | 21166 | -1.348 | -0.2550 | Yes | ||
| 100 | ZBTB9 | 1429125_at | 21296 | -1.457 | -0.2425 | Yes | ||
| 101 | GPHN | 1426462_at 1426463_at 1430038_at 1444869_at 1445188_at 1458242_at | 21518 | -1.682 | -0.2313 | Yes | ||
| 102 | LTA | 1420353_at | 21540 | -1.710 | -0.2105 | Yes | ||
| 103 | CPLX2 | 1421477_at 1436383_at 1455672_s_at | 21606 | -1.836 | -0.1902 | Yes | ||
| 104 | IRF1 | 1448436_a_at | 21686 | -2.021 | -0.1682 | Yes | ||
| 105 | ZHX2 | 1455210_at | 21747 | -2.244 | -0.1425 | Yes | ||
| 106 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 21839 | -2.911 | -0.1098 | Yes | ||
| 107 | NR4A1 | 1416505_at | 21917 | -3.996 | -0.0626 | Yes | ||
| 108 | EGR1 | 1417065_at | 21922 | -4.993 | 0.0005 | Yes |