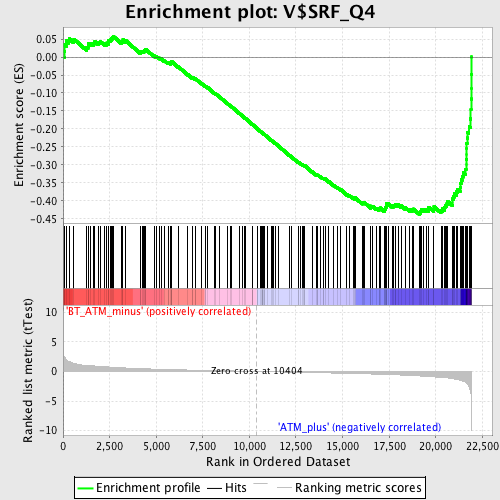
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$SRF_Q4 |
| Enrichment Score (ES) | -0.43785334 |
| Normalized Enrichment Score (NES) | -1.8141991 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.023527164 |
| FWER p-Value | 0.071 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AGRP | 1421690_s_at 1457362_at | 57 | 2.476 | 0.0164 | No | ||
| 2 | CITED2 | 1417129_a_at 1421267_a_at 1439915_at 1440091_at 1443561_at 1443926_at 1445638_at 1446176_at 1447861_x_at 1457632_s_at 1459301_at | 73 | 2.425 | 0.0344 | No | ||
| 3 | ROCK2 | 1423592_at 1451041_at | 175 | 1.980 | 0.0450 | No | ||
| 4 | EML4 | 1429612_at 1447838_x_at | 316 | 1.655 | 0.0513 | No | ||
| 5 | NR2F2 | 1416158_at 1416159_at 1416160_at 1431237_at 1444229_at | 578 | 1.356 | 0.0497 | No | ||
| 6 | FOXP1 | 1421140_a_at 1421141_a_at 1421142_s_at 1435222_at 1438802_at 1443258_at 1446280_at 1447209_at 1455242_at 1456851_at | 1251 | 1.019 | 0.0267 | No | ||
| 7 | PRM1 | 1415955_x_at 1437054_x_at 1439379_x_at | 1339 | 1.001 | 0.0304 | No | ||
| 8 | RASD2 | 1427343_at 1427344_s_at 1458433_at | 1358 | 0.999 | 0.0373 | No | ||
| 9 | ARHGEF17 | 1419938_s_at 1419939_at 1433682_at 1442357_at 1447441_at | 1488 | 0.972 | 0.0388 | No | ||
| 10 | TNMD | 1417979_at | 1647 | 0.937 | 0.0388 | No | ||
| 11 | VCL | 1416156_at 1416157_at 1445256_at | 1706 | 0.924 | 0.0432 | No | ||
| 12 | PPP1R12A | 1429487_at 1437734_at 1437735_at 1444762_at 1444834_at 1453163_at | 1909 | 0.877 | 0.0407 | No | ||
| 13 | STAG2 | 1421849_at 1450396_at | 1991 | 0.857 | 0.0435 | No | ||
| 14 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 2233 | 0.801 | 0.0386 | No | ||
| 15 | RARB | 1454906_at | 2355 | 0.772 | 0.0390 | No | ||
| 16 | POU3F4 | 1422164_at 1422165_at | 2436 | 0.757 | 0.0412 | No | ||
| 17 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 2454 | 0.753 | 0.0462 | No | ||
| 18 | SCOC | 1416267_at 1430999_a_at | 2546 | 0.731 | 0.0476 | No | ||
| 19 | RHOJ | 1418892_at 1444982_at | 2586 | 0.723 | 0.0514 | No | ||
| 20 | MAP2K6 | 1426850_a_at 1459292_at | 2630 | 0.715 | 0.0549 | No | ||
| 21 | EMILIN2 | 1435264_at | 2679 | 0.704 | 0.0581 | No | ||
| 22 | ATRX | 1420945_at 1420946_at 1420947_at 1438750_at 1441014_at 1441771_at 1450051_at 1453734_at 1456832_at | 3151 | 0.619 | 0.0412 | No | ||
| 23 | VGF | 1436094_at | 3199 | 0.611 | 0.0438 | No | ||
| 24 | MMP14 | 1416572_at 1448383_at | 3213 | 0.608 | 0.0479 | No | ||
| 25 | SLC35A2 | 1422827_x_at 1432533_a_at 1436664_a_at 1439433_a_at 1453858_at 1457024_x_at | 3364 | 0.584 | 0.0455 | No | ||
| 26 | DVL3 | 1420457_at | 4143 | 0.479 | 0.0134 | No | ||
| 27 | TBL1X | 1434643_at 1434644_at 1441435_at 1446736_at 1455042_at | 4158 | 0.478 | 0.0164 | No | ||
| 28 | TGFB1I1 | 1418136_at | 4236 | 0.468 | 0.0165 | No | ||
| 29 | CTGF | 1416953_at | 4313 | 0.461 | 0.0166 | No | ||
| 30 | JMJD1A | 1426810_at | 4371 | 0.455 | 0.0174 | No | ||
| 31 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 4404 | 0.451 | 0.0194 | No | ||
| 32 | RIMS1 | 1435667_at 1438305_at 1444393_at | 4420 | 0.449 | 0.0222 | No | ||
| 33 | PPP2R3A | 1455198_a_at | 4903 | 0.400 | 0.0031 | No | ||
| 34 | TANK | 1421640_a_at 1441513_at 1459668_at | 5024 | 0.388 | 0.0006 | No | ||
| 35 | TPM2 | 1419738_a_at 1419739_at 1425028_a_at 1449577_x_at | 5160 | 0.375 | -0.0027 | No | ||
| 36 | ASPH | 1420959_at 1425274_at 1425275_at 1426015_s_at 1436455_at 1450058_at | 5256 | 0.366 | -0.0043 | No | ||
| 37 | PODN | 1435180_at | 5457 | 0.347 | -0.0108 | No | ||
| 38 | ITGB1BP2 | 1423238_at | 5641 | 0.332 | -0.0166 | No | ||
| 39 | CSRP1 | 1425810_a_at 1425811_a_at | 5649 | 0.331 | -0.0144 | No | ||
| 40 | CKM | 1417614_at | 5769 | 0.321 | -0.0174 | No | ||
| 41 | PPAP2B | 1429514_at 1446850_at 1448908_at | 5797 | 0.318 | -0.0162 | No | ||
| 42 | MYL1 | 1452651_a_at | 5811 | 0.316 | -0.0144 | No | ||
| 43 | SH3BGR | 1422644_at | 5812 | 0.316 | -0.0119 | No | ||
| 44 | HERC1 | 1425378_at 1434060_at 1440437_at 1440903_at | 6184 | 0.285 | -0.0268 | No | ||
| 45 | TADA3L | 1417467_a_at 1448719_at | 6696 | 0.247 | -0.0484 | No | ||
| 46 | SRF | 1418255_s_at 1418256_at | 6937 | 0.228 | -0.0577 | No | ||
| 47 | SDPR | 1416778_at 1416779_at 1443832_s_at | 6971 | 0.225 | -0.0575 | No | ||
| 48 | HOXB5 | 1418415_at | 7108 | 0.215 | -0.0620 | No | ||
| 49 | SRD5A2 | 1422960_at | 7114 | 0.215 | -0.0606 | No | ||
| 50 | CALN1 | 1425846_a_at | 7432 | 0.193 | -0.0737 | No | ||
| 51 | FGF17 | 1421523_at 1456239_at | 7632 | 0.179 | -0.0815 | No | ||
| 52 | CHD2 | 1444246_at 1445843_at | 7648 | 0.178 | -0.0808 | No | ||
| 53 | EFHD1 | 1448507_at | 7770 | 0.170 | -0.0851 | No | ||
| 54 | MYL9 | 1452670_at | 8147 | 0.143 | -0.1012 | No | ||
| 55 | PPARG | 1420715_a_at | 8208 | 0.139 | -0.1029 | No | ||
| 56 | MYLK | 1425504_at 1425505_at 1425506_at | 8406 | 0.126 | -0.1110 | No | ||
| 57 | CFL2 | 1418066_at 1418067_at 1431432_at | 8810 | 0.101 | -0.1287 | No | ||
| 58 | DSTN | 1417124_at | 9007 | 0.089 | -0.1371 | No | ||
| 59 | NR2F1 | 1418157_at 1422412_x_at | 9033 | 0.087 | -0.1375 | No | ||
| 60 | CDKN1B | 1419497_at 1434045_at | 9462 | 0.061 | -0.1567 | No | ||
| 61 | CSDA | 1435800_a_at 1451012_a_at | 9619 | 0.050 | -0.1635 | No | ||
| 62 | TMOD1 | 1419863_at 1422754_at | 9761 | 0.042 | -0.1697 | No | ||
| 63 | NPR3 | 1435184_at 1448024_at 1450286_at | 9789 | 0.040 | -0.1706 | No | ||
| 64 | SERPINH1 | 1450843_a_at 1456733_x_at | 10147 | 0.016 | -0.1869 | No | ||
| 65 | EGR4 | 1449977_at | 10167 | 0.015 | -0.1877 | No | ||
| 66 | TAGLN | 1423505_at | 10169 | 0.015 | -0.1876 | No | ||
| 67 | GAB2 | 1419829_a_at 1420785_at 1439786_at 1446860_at | 10443 | -0.002 | -0.2001 | No | ||
| 68 | MYH11 | 1418122_at 1448962_at | 10445 | -0.002 | -0.2002 | No | ||
| 69 | ELF4 | 1421337_at 1445778_at | 10624 | -0.013 | -0.2082 | No | ||
| 70 | MRGPRF | 1425894_at | 10677 | -0.016 | -0.2105 | No | ||
| 71 | KCNMB1 | 1421400_at 1421401_at | 10696 | -0.017 | -0.2112 | No | ||
| 72 | ADAMTSL1 | 1430313_at | 10701 | -0.018 | -0.2113 | No | ||
| 73 | IGF2AS | 1427394_at | 10720 | -0.018 | -0.2119 | No | ||
| 74 | MUS81 | 1431885_a_at 1450432_s_at | 10736 | -0.019 | -0.2125 | No | ||
| 75 | CRH | 1457984_at | 10788 | -0.023 | -0.2146 | No | ||
| 76 | TRIM46 | 1460568_at | 10824 | -0.026 | -0.2161 | No | ||
| 77 | TNNC1 | 1418370_at | 10996 | -0.038 | -0.2236 | No | ||
| 78 | KCNK3 | 1425341_at 1425342_a_at 1426058_a_at | 11167 | -0.048 | -0.2311 | No | ||
| 79 | AKAP12 | 1419706_a_at 1458235_at | 11231 | -0.053 | -0.2336 | No | ||
| 80 | BNC2 | 1438861_at 1440593_at 1459132_at | 11271 | -0.055 | -0.2349 | No | ||
| 81 | DGKG | 1419756_at 1431167_at 1449584_at | 11415 | -0.063 | -0.2410 | No | ||
| 82 | TAF5L | 1448195_at | 11574 | -0.072 | -0.2477 | No | ||
| 83 | SRP54 | 1430476_at 1441233_at 1441480_at | 12154 | -0.108 | -0.2735 | No | ||
| 84 | PLCB3 | 1448661_at | 12247 | -0.113 | -0.2769 | No | ||
| 85 | CACNA1B | 1425812_a_at 1436602_x_at 1439612_at 1460608_at | 12619 | -0.137 | -0.2929 | No | ||
| 86 | ASPA | 1418472_at | 12647 | -0.138 | -0.2930 | No | ||
| 87 | TAZ | 1416207_at 1453785_at | 12737 | -0.145 | -0.2960 | No | ||
| 88 | MID1 | 1422055_at 1438239_at 1441929_at 1443666_at | 12832 | -0.151 | -0.2992 | No | ||
| 89 | NLGN3 | 1456384_at | 12897 | -0.155 | -0.3009 | No | ||
| 90 | EDN1 | 1451924_a_at | 12943 | -0.158 | -0.3018 | No | ||
| 91 | FGFRL1 | 1447878_s_at 1451912_a_at | 12979 | -0.160 | -0.3021 | No | ||
| 92 | NKX2-2 | 1421112_at | 13380 | -0.186 | -0.3191 | No | ||
| 93 | FOXP2 | 1422014_at 1438231_at 1438232_at 1440108_at 1441365_at 1458191_at | 13585 | -0.200 | -0.3269 | No | ||
| 94 | H2AFX | 1416746_at | 13647 | -0.204 | -0.3282 | No | ||
| 95 | MSX1 | 1417127_at 1448601_s_at | 13678 | -0.207 | -0.3280 | No | ||
| 96 | NPAS2 | 1421036_at 1421037_at | 13829 | -0.217 | -0.3332 | No | ||
| 97 | BLOC1S1 | 1422614_s_at | 13959 | -0.225 | -0.3374 | No | ||
| 98 | PFN1 | 1449018_at | 13989 | -0.227 | -0.3370 | No | ||
| 99 | FOSB | 1422134_at | 14066 | -0.232 | -0.3387 | No | ||
| 100 | CNN1 | 1417917_at | 14273 | -0.247 | -0.3462 | No | ||
| 101 | FOXE3 | 1422393_at | 14544 | -0.266 | -0.3566 | No | ||
| 102 | SORT1 | 1423362_at 1423363_at 1450955_s_at | 14719 | -0.280 | -0.3624 | No | ||
| 103 | RBBP7 | 1415775_at 1456227_x_at | 14920 | -0.294 | -0.3694 | No | ||
| 104 | HOXD10 | 1418606_at | 15240 | -0.318 | -0.3816 | No | ||
| 105 | ESRRA | 1442864_at 1460652_at | 15372 | -0.326 | -0.3851 | No | ||
| 106 | MYL7 | 1449071_at | 15574 | -0.342 | -0.3917 | No | ||
| 107 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 15670 | -0.350 | -0.3934 | No | ||
| 108 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 15686 | -0.351 | -0.3914 | No | ||
| 109 | KRTCAP2 | 1417058_a_at 1417059_at | 16068 | -0.382 | -0.4059 | No | ||
| 110 | MAP1A | 1460566_at | 16104 | -0.386 | -0.4046 | No | ||
| 111 | MAPKAPK2 | 1426648_at | 16184 | -0.392 | -0.4052 | No | ||
| 112 | PHF12 | 1434922_at 1453579_at | 16518 | -0.422 | -0.4172 | No | ||
| 113 | IL17B | 1431693_a_at | 16523 | -0.423 | -0.4142 | No | ||
| 114 | ABR | 1433477_at | 16610 | -0.430 | -0.4148 | No | ||
| 115 | LRRTM4 | 1437214_at 1455937_at | 16826 | -0.451 | -0.4212 | No | ||
| 116 | DLL1 | 1419204_at | 16966 | -0.465 | -0.4240 | No | ||
| 117 | ARPC4 | 1423588_at 1423589_at | 16995 | -0.468 | -0.4217 | No | ||
| 118 | INSM1 | 1421399_at 1455865_at | 17027 | -0.471 | -0.4195 | No | ||
| 119 | JPH2 | 1421453_at 1455404_at | 17244 | -0.494 | -0.4257 | No | ||
| 120 | IHPK1 | 1422968_at 1422969_s_at 1438845_at 1438846_x_at 1452694_at | 17269 | -0.496 | -0.4229 | No | ||
| 121 | TGIF | 1422286_a_at | 17305 | -0.499 | -0.4207 | No | ||
| 122 | MYO1E | 1420159_at 1420160_s_at 1428509_at 1449753_at 1449941_at | 17324 | -0.502 | -0.4177 | No | ||
| 123 | SERTAD4 | 1454877_at | 17367 | -0.508 | -0.4157 | No | ||
| 124 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 17373 | -0.508 | -0.4120 | No | ||
| 125 | NPPA | 1456062_at | 17376 | -0.509 | -0.4082 | No | ||
| 126 | GRK6 | 1437436_s_at 1450480_a_at 1451672_at | 17448 | -0.516 | -0.4075 | No | ||
| 127 | SLC4A3 | 1418485_at 1453687_at | 17682 | -0.543 | -0.4140 | No | ||
| 128 | MYL6 | 1435041_at | 17753 | -0.551 | -0.4130 | No | ||
| 129 | HOXB4 | 1451761_at 1460379_at | 17824 | -0.559 | -0.4119 | No | ||
| 130 | HOXC5 | 1439885_at 1450832_at 1456301_at | 17858 | -0.564 | -0.4091 | No | ||
| 131 | ACTG2 | 1422340_a_at | 17989 | -0.581 | -0.4106 | No | ||
| 132 | CFL1 | 1448346_at 1455138_x_at | 18162 | -0.606 | -0.4138 | No | ||
| 133 | EEF1B2 | 1448252_a_at | 18361 | -0.631 | -0.4181 | No | ||
| 134 | RSU1 | 1419259_at | 18613 | -0.669 | -0.4245 | No | ||
| 135 | THRAP1 | 1436904_at 1438725_at 1444068_at 1446840_at 1453160_at 1453161_at | 18741 | -0.691 | -0.4250 | No | ||
| 136 | DUSP5 | 1437199_at | 18806 | -0.699 | -0.4225 | No | ||
| 137 | MCL1 | 1416880_at 1416881_at 1437527_x_at 1448503_at 1456243_x_at 1456381_x_at | 19140 | -0.753 | -0.4321 | Yes | ||
| 138 | HOXA5 | 1443803_x_at 1448926_at | 19187 | -0.762 | -0.4283 | Yes | ||
| 139 | FGF8 | 1451882_a_at | 19222 | -0.771 | -0.4239 | Yes | ||
| 140 | TPM1 | 1423049_a_at 1423721_at 1447713_at 1456623_at | 19366 | -0.796 | -0.4244 | Yes | ||
| 141 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 19502 | -0.822 | -0.4243 | Yes | ||
| 142 | RAB30 | 1426452_a_at | 19627 | -0.847 | -0.4235 | Yes | ||
| 143 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 19632 | -0.847 | -0.4171 | Yes | ||
| 144 | GPR20 | 1440021_at | 19906 | -0.909 | -0.4227 | Yes | ||
| 145 | EGR3 | 1421486_at 1436329_at | 19914 | -0.912 | -0.4160 | Yes | ||
| 146 | RUNDC1 | 1454829_at | 20295 | -1.003 | -0.4257 | Yes | ||
| 147 | ACTA1 | 1427735_a_at | 20375 | -1.018 | -0.4215 | Yes | ||
| 148 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20490 | -1.047 | -0.4187 | Yes | ||
| 149 | TLE3 | 1419654_at 1419655_at 1449554_at 1458512_at | 20515 | -1.053 | -0.4117 | Yes | ||
| 150 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 20612 | -1.081 | -0.4078 | Yes | ||
| 151 | ERBB2IP | 1428011_a_at 1439079_a_at 1439080_at 1439638_at 1440573_at | 20654 | -1.094 | -0.4013 | Yes | ||
| 152 | FST | 1421365_at 1434458_at | 20913 | -1.213 | -0.4038 | Yes | ||
| 153 | KCNA1 | 1417416_at 1437230_at 1442413_at 1455785_at | 20914 | -1.213 | -0.3944 | Yes | ||
| 154 | STAT5B | 1422102_a_at 1422103_a_at | 20958 | -1.236 | -0.3869 | Yes | ||
| 155 | MYO1B | 1427450_x_at 1441210_at 1447364_x_at 1448989_a_at 1448990_a_at 1458007_at 1459679_s_at | 21013 | -1.260 | -0.3797 | Yes | ||
| 156 | DIXDC1 | 1425256_a_at 1435207_at 1443957_at 1444395_at | 21126 | -1.322 | -0.3747 | Yes | ||
| 157 | GADD45G | 1453851_a_at | 21192 | -1.365 | -0.3672 | Yes | ||
| 158 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 21320 | -1.472 | -0.3617 | Yes | ||
| 159 | NCOA6 | 1423374_at | 21331 | -1.486 | -0.3507 | Yes | ||
| 160 | SGK | 1416041_at | 21376 | -1.525 | -0.3410 | Yes | ||
| 161 | COL23A1 | 1429209_at 1429210_at 1440911_at | 21427 | -1.586 | -0.3311 | Yes | ||
| 162 | SVIL | 1427662_at 1460694_s_at 1460735_at | 21485 | -1.643 | -0.3210 | Yes | ||
| 163 | TPM3 | 1427260_a_at 1427567_a_at 1436958_x_at 1449996_a_at 1449997_at | 21616 | -1.853 | -0.3128 | Yes | ||
| 164 | RERE | 1454670_at | 21641 | -1.906 | -0.2992 | Yes | ||
| 165 | VASP | 1451097_at | 21643 | -1.914 | -0.2845 | Yes | ||
| 166 | FLNA | 1426677_at | 21670 | -1.984 | -0.2704 | Yes | ||
| 167 | JUNB | 1415899_at | 21672 | -1.992 | -0.2552 | Yes | ||
| 168 | ANXA6 | 1415818_at 1429246_a_at 1429247_at 1442288_at | 21677 | -2.002 | -0.2399 | Yes | ||
| 169 | DUSP2 | 1450698_at | 21696 | -2.054 | -0.2250 | Yes | ||
| 170 | RASSF2 | 1428392_at 1444889_at | 21715 | -2.124 | -0.2094 | Yes | ||
| 171 | LPP | 1425673_at 1436714_at 1437235_x_at 1438271_at 1440167_s_at 1454899_at 1455314_at | 21796 | -2.538 | -0.1936 | Yes | ||
| 172 | IER2 | 1416442_at | 21874 | -3.262 | -0.1720 | Yes | ||
| 173 | FOS | 1423100_at | 21901 | -3.644 | -0.1452 | Yes | ||
| 174 | DUSP6 | 1415834_at | 21907 | -3.749 | -0.1165 | Yes | ||
| 175 | NR4A1 | 1416505_at | 21917 | -3.996 | -0.0862 | Yes | ||
| 176 | EGR1 | 1417065_at | 21922 | -4.993 | -0.0480 | Yes | ||
| 177 | EGR2 | 1427682_a_at 1427683_at | 21930 | -6.298 | 0.0002 | Yes |