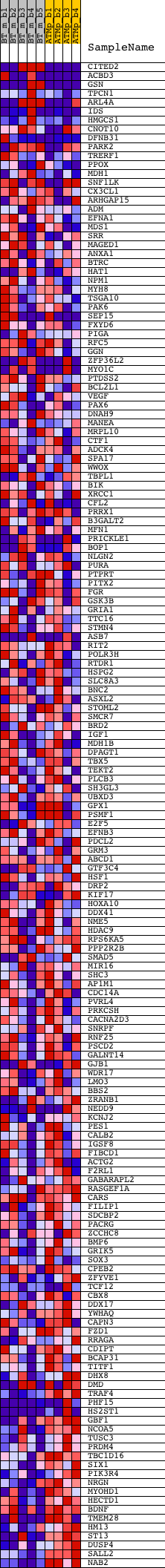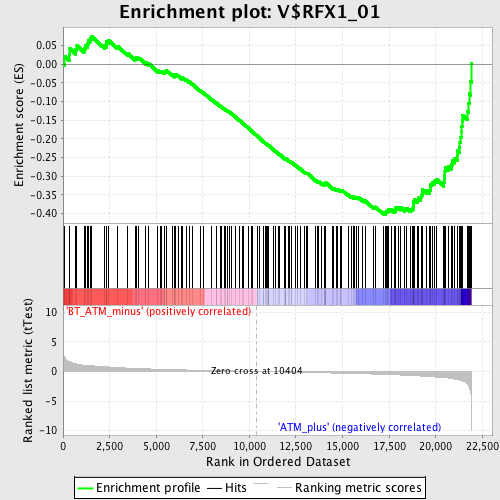
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$RFX1_01 |
| Enrichment Score (ES) | -0.4022336 |
| Normalized Enrichment Score (NES) | -1.6756175 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.038941216 |
| FWER p-Value | 0.369 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CITED2 | 1417129_a_at 1421267_a_at 1439915_at 1440091_at 1443561_at 1443926_at 1445638_at 1446176_at 1447861_x_at 1457632_s_at 1459301_at | 73 | 2.425 | 0.0210 | No | ||
| 2 | ACBD3 | 1426635_at 1428668_at 1452137_at 1456316_a_at | 321 | 1.650 | 0.0263 | No | ||
| 3 | GSN | 1415812_at 1436991_x_at 1437171_x_at 1441225_at 1456312_x_at 1456568_at 1456569_x_at | 353 | 1.622 | 0.0412 | No | ||
| 4 | TPCN1 | 1434930_at 1451771_at 1451772_at | 672 | 1.287 | 0.0395 | No | ||
| 5 | ARL4A | 1425411_at 1431429_a_at 1435092_at | 741 | 1.246 | 0.0489 | No | ||
| 6 | IDS | 1421216_a_at 1427812_at 1427813_at 1434751_at 1450166_at 1455481_at | 1126 | 1.051 | 0.0418 | No | ||
| 7 | HMGCS1 | 1433443_a_at 1433444_at 1433445_x_at 1433446_at 1441536_at | 1196 | 1.033 | 0.0490 | No | ||
| 8 | CNOT10 | 1433882_at 1453654_at 1459756_at 1459757_x_at | 1302 | 1.010 | 0.0544 | No | ||
| 9 | DFNB31 | 1432555_at 1436485_s_at 1436486_x_at 1442507_at 1447438_at | 1350 | 1.000 | 0.0623 | No | ||
| 10 | PARK2 | 1420755_a_at 1426135_a_at 1449975_a_at | 1445 | 0.980 | 0.0678 | No | ||
| 11 | TRERF1 | 1444654_at | 1507 | 0.967 | 0.0747 | No | ||
| 12 | PPOX | 1416618_at 1441862_at 1448065_at | 2210 | 0.806 | 0.0506 | No | ||
| 13 | MDH1 | 1434319_at 1436834_x_at 1438338_at 1448172_at 1454925_x_at | 2326 | 0.781 | 0.0531 | No | ||
| 14 | SNF1LK | 1419766_at 1459601_at | 2335 | 0.779 | 0.0606 | No | ||
| 15 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 2454 | 0.753 | 0.0627 | No | ||
| 16 | ARHGAP15 | 1429881_at 1435959_at 1441650_at | 2927 | 0.660 | 0.0477 | No | ||
| 17 | ADM | 1416077_at 1447839_x_at | 3456 | 0.569 | 0.0292 | No | ||
| 18 | EFNA1 | 1416895_at 1448510_at | 3861 | 0.514 | 0.0158 | No | ||
| 19 | MDS1 | 1422295_at | 3917 | 0.508 | 0.0183 | No | ||
| 20 | SRR | 1419664_at 1438549_a_at 1438550_x_at 1453421_at 1455045_at 1458393_at | 4071 | 0.489 | 0.0162 | No | ||
| 21 | MAGED1 | 1450062_a_at | 4433 | 0.447 | 0.0041 | No | ||
| 22 | ANXA1 | 1448213_at | 4607 | 0.429 | 0.0005 | No | ||
| 23 | BTRC | 1417325_at 1425680_a_at 1443142_at 1446925_at 1447120_at 1456780_at | 5087 | 0.382 | -0.0177 | No | ||
| 24 | HAT1 | 1428061_at | 5218 | 0.370 | -0.0199 | No | ||
| 25 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 5290 | 0.363 | -0.0195 | No | ||
| 26 | MYH8 | 1426650_at | 5434 | 0.349 | -0.0226 | No | ||
| 27 | TSGA10 | 1436045_at 1447283_at | 5470 | 0.347 | -0.0207 | No | ||
| 28 | PAK6 | 1445475_at 1455200_at | 5533 | 0.342 | -0.0201 | No | ||
| 29 | SEP15 | 1448903_at 1454525_at | 5548 | 0.340 | -0.0173 | No | ||
| 30 | FXYD6 | 1417343_at | 5856 | 0.311 | -0.0283 | No | ||
| 31 | PIGA | 1427305_at 1453497_a_at | 5995 | 0.301 | -0.0316 | No | ||
| 32 | RFC5 | 1452917_at | 6006 | 0.299 | -0.0291 | No | ||
| 33 | GGN | 1445278_at | 6058 | 0.295 | -0.0285 | No | ||
| 34 | ZFP36L2 | 1437626_at 1439054_at 1451034_at | 6185 | 0.285 | -0.0314 | No | ||
| 35 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 6378 | 0.271 | -0.0375 | No | ||
| 36 | PTDSS2 | 1426728_x_at 1429492_x_at 1434586_a_at 1434587_x_at 1439984_at 1443738_at 1450354_a_at 1453164_a_at 1458387_at 1460010_a_at | 6397 | 0.269 | -0.0356 | No | ||
| 37 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 6603 | 0.252 | -0.0425 | No | ||
| 38 | VEGF | 1420909_at 1451959_a_at | 6760 | 0.242 | -0.0472 | No | ||
| 39 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 6943 | 0.227 | -0.0533 | No | ||
| 40 | DNAH9 | 1442412_at 1457401_at | 7354 | 0.198 | -0.0701 | No | ||
| 41 | MANEA | 1455403_at | 7523 | 0.187 | -0.0760 | No | ||
| 42 | MRPL10 | 1418112_at | 7986 | 0.155 | -0.0956 | No | ||
| 43 | CTF1 | 1449093_at | 8225 | 0.138 | -0.1052 | No | ||
| 44 | ADCK4 | 1448490_at | 8228 | 0.138 | -0.1039 | No | ||
| 45 | SPA17 | 1417635_at | 8433 | 0.124 | -0.1120 | No | ||
| 46 | WWOX | 1416334_at 1431942_at 1431960_at 1438517_at 1442651_at 1446412_at | 8497 | 0.120 | -0.1137 | No | ||
| 47 | TBPL1 | 1431415_a_at 1434907_at | 8644 | 0.111 | -0.1193 | No | ||
| 48 | BIK | 1420362_a_at 1420363_at 1449836_x_at | 8733 | 0.105 | -0.1223 | No | ||
| 49 | XRCC1 | 1416587_a_at | 8808 | 0.101 | -0.1247 | No | ||
| 50 | CFL2 | 1418066_at 1418067_at 1431432_at | 8810 | 0.101 | -0.1237 | No | ||
| 51 | PRRX1 | 1425526_a_at 1425527_at 1425528_at 1432129_a_at 1439774_at 1458004_at | 8921 | 0.094 | -0.1278 | No | ||
| 52 | B3GALT2 | 1423084_at 1437433_at 1437644_at | 9061 | 0.086 | -0.1333 | No | ||
| 53 | MFN1 | 1426773_at 1449697_s_at | 9235 | 0.075 | -0.1405 | No | ||
| 54 | PRICKLE1 | 1441663_at 1442172_at 1442400_at 1444759_at 1452249_at 1457637_at | 9454 | 0.061 | -0.1499 | No | ||
| 55 | BOP1 | 1423264_at 1430491_at | 9645 | 0.049 | -0.1582 | No | ||
| 56 | NLGN2 | 1455143_at 1460013_at | 9692 | 0.046 | -0.1598 | No | ||
| 57 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 9977 | 0.027 | -0.1726 | No | ||
| 58 | PTPRT | 1421238_a_at 1439725_at 1447135_at 1450174_at | 10093 | 0.019 | -0.1777 | No | ||
| 59 | PITX2 | 1424797_a_at 1450482_a_at | 10173 | 0.014 | -0.1812 | No | ||
| 60 | FGR | 1419526_at 1442804_at | 10416 | -0.000 | -0.1923 | No | ||
| 61 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 10461 | -0.003 | -0.1943 | No | ||
| 62 | GRIA1 | 1435239_at 1448972_at 1458285_at | 10538 | -0.008 | -0.1977 | No | ||
| 63 | TTC16 | 1455414_at | 10750 | -0.021 | -0.2072 | No | ||
| 64 | STMN4 | 1418105_at | 10758 | -0.021 | -0.2073 | No | ||
| 65 | ASB7 | 1435010_at 1438744_at 1446319_at 1450363_at | 10887 | -0.031 | -0.2129 | No | ||
| 66 | RIT2 | 1421779_at 1448125_at 1460166_at | 10946 | -0.035 | -0.2152 | No | ||
| 67 | POLR3H | 1424227_at 1424228_at | 10954 | -0.035 | -0.2151 | No | ||
| 68 | RTDR1 | 1431601_at | 11015 | -0.039 | -0.2175 | No | ||
| 69 | HSPG2 | 1418669_at 1418670_s_at | 11021 | -0.040 | -0.2173 | No | ||
| 70 | SLC8A3 | 1440962_at 1450311_at | 11035 | -0.041 | -0.2175 | No | ||
| 71 | BNC2 | 1438861_at 1440593_at 1459132_at | 11271 | -0.055 | -0.2278 | No | ||
| 72 | ASXL2 | 1439063_at 1444754_at 1453295_at 1458366_at 1460597_at | 11412 | -0.063 | -0.2336 | No | ||
| 73 | STOML2 | 1448774_at | 11585 | -0.073 | -0.2407 | No | ||
| 74 | SMCR7 | 1434072_at | 11645 | -0.077 | -0.2427 | No | ||
| 75 | BRD2 | 1423502_at 1437210_a_at | 11867 | -0.090 | -0.2519 | No | ||
| 76 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 11916 | -0.094 | -0.2532 | No | ||
| 77 | MDH1B | 1429842_at | 11917 | -0.094 | -0.2522 | No | ||
| 78 | DPAGT1 | 1448549_a_at | 11950 | -0.096 | -0.2527 | No | ||
| 79 | TBX5 | 1425694_at 1425695_at | 12080 | -0.103 | -0.2576 | No | ||
| 80 | TEKT2 | 1418484_at | 12161 | -0.108 | -0.2602 | No | ||
| 81 | PLCB3 | 1448661_at | 12247 | -0.113 | -0.2630 | No | ||
| 82 | SH3GL3 | 1432103_a_at | 12281 | -0.115 | -0.2633 | No | ||
| 83 | UBXD3 | 1426007_a_at 1451906_at 1456385_x_at 1457581_at | 12453 | -0.127 | -0.2699 | No | ||
| 84 | GPX1 | 1460671_at | 12601 | -0.136 | -0.2753 | No | ||
| 85 | PSMF1 | 1424369_at 1424370_s_at 1424371_at 1451337_at | 12740 | -0.145 | -0.2802 | No | ||
| 86 | E2F5 | 1417444_at 1447625_at 1460207_s_at | 12973 | -0.159 | -0.2892 | No | ||
| 87 | EFNB3 | 1423085_at | 13056 | -0.164 | -0.2914 | No | ||
| 88 | PDCL2 | 1432503_a_at | 13107 | -0.168 | -0.2920 | No | ||
| 89 | GRM3 | 1430136_at | 13566 | -0.198 | -0.3110 | No | ||
| 90 | ABCD1 | 1418838_at | 13684 | -0.207 | -0.3143 | No | ||
| 91 | GTF3C4 | 1434320_at 1438692_at | 13703 | -0.208 | -0.3130 | No | ||
| 92 | HSF1 | 1424622_at | 13898 | -0.221 | -0.3197 | No | ||
| 93 | DRP2 | 1440452_at 1441224_at 1441542_at | 14012 | -0.228 | -0.3226 | No | ||
| 94 | KIF17 | 1419826_at 1419827_s_at 1422762_at | 14056 | -0.232 | -0.3223 | No | ||
| 95 | HOXA10 | 1431475_a_at 1446408_at | 14080 | -0.233 | -0.3210 | No | ||
| 96 | DDX41 | 1423814_at | 14085 | -0.234 | -0.3188 | No | ||
| 97 | NME5 | 1422727_at | 14114 | -0.236 | -0.3177 | No | ||
| 98 | HDAC9 | 1421306_a_at 1434572_at | 14480 | -0.261 | -0.3319 | No | ||
| 99 | RPS6KA5 | 1431050_at 1437625_at 1439004_at 1440343_at | 14533 | -0.265 | -0.3316 | No | ||
| 100 | PPP2R2B | 1426621_a_at | 14684 | -0.278 | -0.3357 | No | ||
| 101 | SMAD5 | 1421047_at 1433641_at 1451873_a_at | 14712 | -0.279 | -0.3341 | No | ||
| 102 | MIR16 | 1418444_a_at | 14871 | -0.290 | -0.3385 | No | ||
| 103 | SHC3 | 1450326_at | 14912 | -0.293 | -0.3374 | No | ||
| 104 | AP1M1 | 1416307_at 1459832_s_at | 14974 | -0.298 | -0.3372 | No | ||
| 105 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 15302 | -0.322 | -0.3490 | No | ||
| 106 | PVRL4 | 1428701_at 1451690_a_at | 15474 | -0.335 | -0.3534 | No | ||
| 107 | PRKCSH | 1416339_a_at | 15610 | -0.344 | -0.3562 | No | ||
| 108 | CACNA2D3 | 1419225_at | 15671 | -0.350 | -0.3554 | No | ||
| 109 | SNRPF | 1428672_at | 15779 | -0.358 | -0.3567 | No | ||
| 110 | RNF25 | 1442108_at 1448674_at | 15870 | -0.366 | -0.3572 | No | ||
| 111 | PSCD2 | 1450103_a_at | 16071 | -0.383 | -0.3625 | No | ||
| 112 | GALNT14 | 1453630_at | 16233 | -0.396 | -0.3659 | No | ||
| 113 | GJB1 | 1448766_at 1448767_s_at | 16687 | -0.437 | -0.3824 | No | ||
| 114 | WDR17 | 1435392_at 1453537_a_at | 16782 | -0.446 | -0.3822 | No | ||
| 115 | LMO3 | 1455754_at | 17198 | -0.489 | -0.3964 | No | ||
| 116 | BBS2 | 1424478_at | 17327 | -0.502 | -0.3972 | Yes | ||
| 117 | ZRANB1 | 1415712_at 1427200_at 1436228_at | 17351 | -0.505 | -0.3932 | Yes | ||
| 118 | NEDD9 | 1422818_at 1437132_x_at 1447885_x_at 1450767_at | 17445 | -0.516 | -0.3923 | Yes | ||
| 119 | KCNJ2 | 1450503_at | 17497 | -0.521 | -0.3894 | Yes | ||
| 120 | PES1 | 1416172_at 1416173_at | 17623 | -0.536 | -0.3897 | Yes | ||
| 121 | CALB2 | 1451129_at | 17791 | -0.555 | -0.3918 | Yes | ||
| 122 | IGSF8 | 1460675_at | 17864 | -0.565 | -0.3894 | Yes | ||
| 123 | FIBCD1 | 1435482_at | 17867 | -0.565 | -0.3838 | Yes | ||
| 124 | ACTG2 | 1422340_a_at | 17989 | -0.581 | -0.3835 | Yes | ||
| 125 | F2RL1 | 1448931_at | 18136 | -0.602 | -0.3842 | Yes | ||
| 126 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 18332 | -0.627 | -0.3869 | Yes | ||
| 127 | RASGEF1A | 1429316_at 1435218_at | 18432 | -0.642 | -0.3850 | Yes | ||
| 128 | CARS | 1427330_at 1452394_at | 18639 | -0.672 | -0.3877 | Yes | ||
| 129 | FILIP1 | 1436650_at 1444075_at | 18749 | -0.692 | -0.3857 | Yes | ||
| 130 | SDCBP2 | 1424090_at | 18794 | -0.697 | -0.3807 | Yes | ||
| 131 | PACRG | 1428751_at | 18820 | -0.700 | -0.3748 | Yes | ||
| 132 | ZCCHC8 | 1423755_at 1441639_at | 18832 | -0.702 | -0.3683 | Yes | ||
| 133 | BMP6 | 1450759_at 1459947_at | 18849 | -0.705 | -0.3619 | Yes | ||
| 134 | GRIK5 | 1418784_at | 19045 | -0.737 | -0.3635 | Yes | ||
| 135 | SOX3 | 1435192_at 1450485_at | 19091 | -0.746 | -0.3580 | Yes | ||
| 136 | CPEB2 | 1434272_at 1443017_at 1458518_at | 19218 | -0.770 | -0.3561 | Yes | ||
| 137 | ZFYVE1 | 1435071_at 1435072_at | 19259 | -0.777 | -0.3501 | Yes | ||
| 138 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 19284 | -0.782 | -0.3433 | Yes | ||
| 139 | CBX8 | 1419198_at 1419199_at | 19304 | -0.785 | -0.3363 | Yes | ||
| 140 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 19502 | -0.822 | -0.3371 | Yes | ||
| 141 | YWHAQ | 1420828_s_at 1420829_a_at 1420830_x_at 1432842_s_at 1437608_x_at 1454378_at 1460590_s_at 1460621_x_at | 19700 | -0.861 | -0.3375 | Yes | ||
| 142 | CAPN3 | 1426043_a_at 1433681_x_at | 19726 | -0.868 | -0.3299 | Yes | ||
| 143 | FZD1 | 1422985_at 1437284_at | 19735 | -0.869 | -0.3215 | Yes | ||
| 144 | RRAGA | 1428905_at | 19815 | -0.887 | -0.3162 | Yes | ||
| 145 | CDIPT | 1423657_at 1436715_s_at | 19938 | -0.918 | -0.3126 | Yes | ||
| 146 | BCAP31 | 1451049_at 1456279_a_at | 20046 | -0.944 | -0.3080 | Yes | ||
| 147 | TITF1 | 1422346_at | 20446 | -1.035 | -0.3160 | Yes | ||
| 148 | DHX8 | 1426629_at | 20481 | -1.045 | -0.3070 | Yes | ||
| 149 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20490 | -1.047 | -0.2969 | Yes | ||
| 150 | TRAF4 | 1416571_at 1460642_at | 20504 | -1.050 | -0.2869 | Yes | ||
| 151 | PHF15 | 1430081_at 1455345_at | 20520 | -1.054 | -0.2770 | Yes | ||
| 152 | HS2ST1 | 1422739_at 1440905_at 1450729_at 1450730_at | 20690 | -1.106 | -0.2736 | Yes | ||
| 153 | GBF1 | 1415709_s_at 1415757_at 1432677_at 1438207_at | 20847 | -1.177 | -0.2690 | Yes | ||
| 154 | NCOA5 | 1424055_at 1456997_at | 20887 | -1.195 | -0.2587 | Yes | ||
| 155 | TUSC3 | 1421662_a_at 1458568_at | 21016 | -1.262 | -0.2519 | Yes | ||
| 156 | PRDM4 | 1452208_at | 21174 | -1.353 | -0.2455 | Yes | ||
| 157 | TBC1D16 | 1435763_at 1442149_at | 21183 | -1.359 | -0.2322 | Yes | ||
| 158 | SIX1 | 1427277_at | 21295 | -1.456 | -0.2227 | Yes | ||
| 159 | PIK3R4 | 1460352_s_at | 21312 | -1.468 | -0.2087 | Yes | ||
| 160 | NRGN | 1423231_at | 21361 | -1.514 | -0.1957 | Yes | ||
| 161 | MYOHD1 | 1451183_at 1459265_at | 21391 | -1.548 | -0.1814 | Yes | ||
| 162 | HECTD1 | 1424141_at 1438039_at | 21395 | -1.553 | -0.1659 | Yes | ||
| 163 | BDNF | 1422168_a_at 1422169_a_at | 21424 | -1.580 | -0.1513 | Yes | ||
| 164 | TMEM28 | 1435138_at 1443271_at 1445869_at 1446912_at 1459483_at | 21452 | -1.604 | -0.1365 | Yes | ||
| 165 | HM13 | 1417287_at 1428855_at 1428856_at 1438456_at 1438865_at 1455631_at | 21735 | -2.196 | -0.1273 | Yes | ||
| 166 | ST13 | 1416851_at 1442775_at 1460193_at | 21782 | -2.418 | -0.1051 | Yes | ||
| 167 | DUSP4 | 1428834_at | 21811 | -2.663 | -0.0796 | Yes | ||
| 168 | SALL2 | 1416638_at | 21892 | -3.581 | -0.0473 | Yes | ||
| 169 | NAB2 | 1417930_at | 21921 | -4.892 | 0.0006 | Yes |