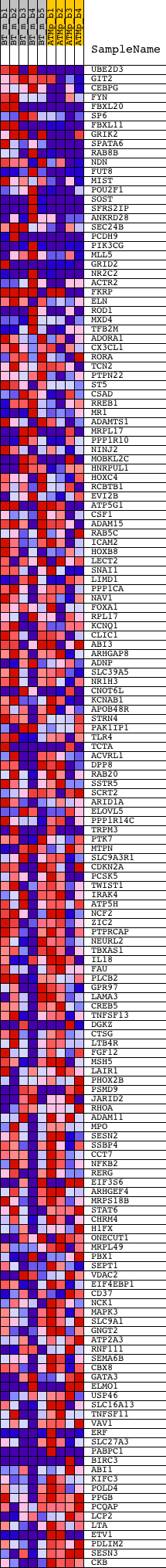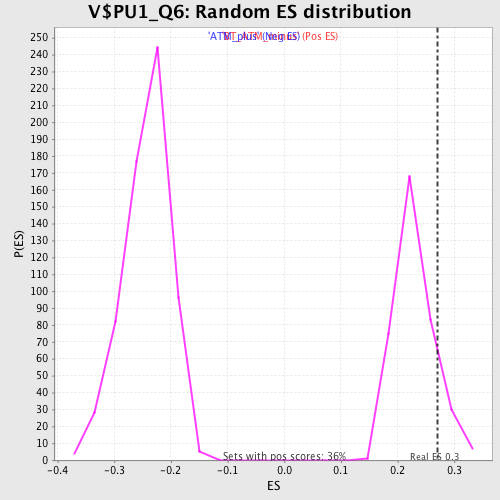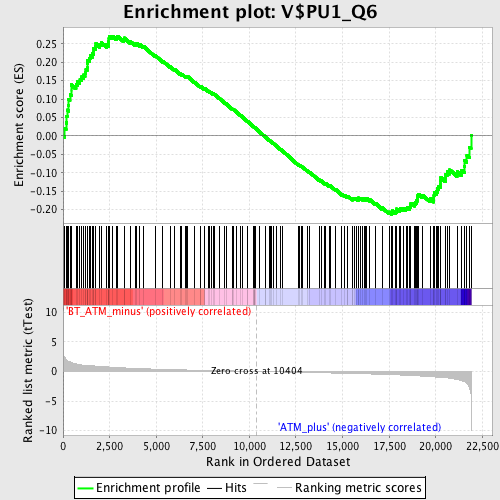
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$PU1_Q6 |
| Enrichment Score (ES) | 0.2709816 |
| Normalized Enrichment Score (NES) | 1.1835531 |
| Nominal p-value | 0.10989011 |
| FDR q-value | 0.58234364 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 77 | 2.421 | 0.0201 | Yes | ||
| 2 | GIT2 | 1423391_at 1432160_at 1435925_at | 158 | 2.034 | 0.0363 | Yes | ||
| 3 | CEBPG | 1425261_at 1425262_at 1451639_at | 196 | 1.906 | 0.0532 | Yes | ||
| 4 | FYN | 1417558_at 1441647_at 1448765_at | 216 | 1.850 | 0.0704 | Yes | ||
| 5 | FBXL20 | 1442464_at 1445575_at 1452826_s_at 1456378_s_at | 293 | 1.686 | 0.0834 | Yes | ||
| 6 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 305 | 1.666 | 0.0992 | Yes | ||
| 7 | FBXL11 | 1435329_at 1443336_at 1444358_at 1455942_at 1458449_at | 370 | 1.610 | 0.1120 | Yes | ||
| 8 | GRIK2 | 1425790_a_at 1439286_at 1440602_at 1445090_at 1457683_at | 438 | 1.544 | 0.1240 | Yes | ||
| 9 | SPATA6 | 1418650_at 1418651_at 1438023_at 1445355_at | 439 | 1.540 | 0.1390 | Yes | ||
| 10 | RAB8B | 1426799_at 1446103_at | 692 | 1.278 | 0.1399 | Yes | ||
| 11 | NDN | 1415923_at 1435382_at 1435383_x_at 1437853_x_at 1438939_x_at 1438978_x_at 1455792_x_at 1456575_at | 781 | 1.219 | 0.1478 | Yes | ||
| 12 | FUT8 | 1441015_at 1442483_at 1443656_at 1456367_at 1460319_at | 905 | 1.142 | 0.1533 | Yes | ||
| 13 | MIST | 1420463_at | 990 | 1.097 | 0.1602 | Yes | ||
| 14 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 1100 | 1.059 | 0.1655 | Yes | ||
| 15 | SOST | 1421245_at 1436240_at 1450179_at | 1177 | 1.037 | 0.1721 | Yes | ||
| 16 | SFRS2IP | 1434691_at 1443085_at 1452884_at 1452885_at 1460445_at | 1226 | 1.025 | 0.1799 | Yes | ||
| 17 | ANKRD28 | 1434630_at 1454801_at | 1288 | 1.012 | 0.1870 | Yes | ||
| 18 | SEC24B | 1426848_at 1426849_at 1458875_at 1459307_at | 1297 | 1.010 | 0.1965 | Yes | ||
| 19 | PCDH9 | 1439322_at 1442659_at 1444215_at 1444724_at 1445052_at 1445061_at 1457973_at | 1315 | 1.007 | 0.2056 | Yes | ||
| 20 | PIK3CG | 1422707_at 1422708_at | 1421 | 0.985 | 0.2104 | Yes | ||
| 21 | MLL5 | 1427236_a_at 1432601_at 1434704_at 1439107_a_at 1439108_at | 1451 | 0.979 | 0.2186 | Yes | ||
| 22 | GRID2 | 1421435_at 1421436_at 1435487_at 1437824_at 1443101_at 1446879_at 1458756_at 1459245_s_at | 1559 | 0.957 | 0.2230 | Yes | ||
| 23 | NR2C2 | 1425014_at 1439696_at 1445613_at 1446955_at 1451569_at 1454167_at 1454851_at | 1620 | 0.943 | 0.2295 | Yes | ||
| 24 | ACTR2 | 1441660_at 1452587_at | 1626 | 0.943 | 0.2385 | Yes | ||
| 25 | FKRP | 1436812_at 1437536_at 1438414_at | 1734 | 0.918 | 0.2425 | Yes | ||
| 26 | ELN | 1420854_at 1420855_at 1446221_at | 1761 | 0.910 | 0.2502 | Yes | ||
| 27 | ROD1 | 1424083_at 1424084_at 1441407_at 1455819_at | 1964 | 0.865 | 0.2494 | Yes | ||
| 28 | MXD4 | 1422574_at 1434378_a_at 1434379_at | 2053 | 0.845 | 0.2536 | Yes | ||
| 29 | TFB2M | 1423441_at | 2332 | 0.779 | 0.2485 | Yes | ||
| 30 | ADORA1 | 1427331_at 1435495_at | 2439 | 0.756 | 0.2510 | Yes | ||
| 31 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 2454 | 0.753 | 0.2577 | Yes | ||
| 32 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 2462 | 0.751 | 0.2647 | Yes | ||
| 33 | TCN2 | 1441850_x_at 1444110_at 1447800_x_at 1447842_x_at 1448200_at 1457825_x_at | 2491 | 0.745 | 0.2707 | Yes | ||
| 34 | PTPN22 | 1417995_at 1442820_at | 2637 | 0.713 | 0.2710 | Yes | ||
| 35 | ST5 | 1428372_at | 2852 | 0.673 | 0.2677 | No | ||
| 36 | CSAD | 1427981_a_at | 2923 | 0.661 | 0.2710 | No | ||
| 37 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 3269 | 0.598 | 0.2610 | No | ||
| 38 | MR1 | 1421898_a_at 1421899_a_at | 3288 | 0.597 | 0.2660 | No | ||
| 39 | ADAMTS1 | 1450716_at | 3629 | 0.546 | 0.2557 | No | ||
| 40 | MRPL17 | 1450866_a_at 1450867_at 1458027_at | 3877 | 0.513 | 0.2494 | No | ||
| 41 | PPP1R10 | 1426726_at 1426727_s_at 1430560_at | 3928 | 0.507 | 0.2520 | No | ||
| 42 | NINJ2 | 1420356_at 1447102_at | 4127 | 0.481 | 0.2476 | No | ||
| 43 | MOBKL2C | 1440919_at 1459919_a_at | 4303 | 0.461 | 0.2441 | No | ||
| 44 | HNRPUL1 | 1451984_at | 4936 | 0.397 | 0.2189 | No | ||
| 45 | HOXC4 | 1422870_at | 5361 | 0.356 | 0.2029 | No | ||
| 46 | RCBTB1 | 1430213_at 1456433_at | 5758 | 0.322 | 0.1879 | No | ||
| 47 | EVI2B | 1426505_at | 5991 | 0.301 | 0.1802 | No | ||
| 48 | ATP5G1 | 1416020_a_at 1444874_at | 6286 | 0.278 | 0.1694 | No | ||
| 49 | CSF1 | 1425154_a_at 1425155_x_at 1448914_a_at 1460220_a_at | 6381 | 0.271 | 0.1677 | No | ||
| 50 | ADAM15 | 1416080_at 1425170_a_at 1438760_x_at 1454206_a_at | 6559 | 0.256 | 0.1621 | No | ||
| 51 | RAB5C | 1424684_at | 6605 | 0.252 | 0.1625 | No | ||
| 52 | ICAM2 | 1448862_at | 6686 | 0.247 | 0.1612 | No | ||
| 53 | HOXB8 | 1452493_s_at | 7058 | 0.218 | 0.1463 | No | ||
| 54 | LECT2 | 1443821_at 1449492_a_at | 7379 | 0.197 | 0.1335 | No | ||
| 55 | SNAI1 | 1448742_at | 7391 | 0.195 | 0.1349 | No | ||
| 56 | LIMD1 | 1422730_at 1422731_at | 7595 | 0.182 | 0.1274 | No | ||
| 57 | PPP1CA | 1460165_at | 7617 | 0.181 | 0.1282 | No | ||
| 58 | NAV1 | 1436907_at 1443629_at 1446497_at | 7789 | 0.169 | 0.1220 | No | ||
| 59 | FOXA1 | 1418496_at | 7870 | 0.163 | 0.1199 | No | ||
| 60 | RPL17 | 1423855_x_at 1435791_x_at 1453752_at | 7966 | 0.156 | 0.1171 | No | ||
| 61 | KCNQ1 | 1441588_at 1442029_at 1446255_at 1447418_at 1449464_at 1457781_at 1459443_at | 8055 | 0.150 | 0.1145 | No | ||
| 62 | CLIC1 | 1416656_at 1458665_at | 8109 | 0.146 | 0.1135 | No | ||
| 63 | ABI3 | 1446191_at 1452928_at | 8395 | 0.127 | 0.1016 | No | ||
| 64 | ARHGAP8 | 1451463_at | 8688 | 0.108 | 0.0893 | No | ||
| 65 | ADNP | 1423069_at | 8747 | 0.104 | 0.0876 | No | ||
| 66 | SLC39A5 | 1429523_a_at | 9083 | 0.084 | 0.0731 | No | ||
| 67 | NR1H3 | 1450444_a_at | 9115 | 0.082 | 0.0725 | No | ||
| 68 | CNOT6L | 1425480_at 1425481_at 1434311_at 1443595_at 1451723_at | 9138 | 0.081 | 0.0722 | No | ||
| 69 | KCNAB1 | 1448468_a_at 1454043_a_at | 9305 | 0.070 | 0.0653 | No | ||
| 70 | APOB48R | 1420382_at | 9529 | 0.057 | 0.0556 | No | ||
| 71 | STRN4 | 1426209_at | 9650 | 0.048 | 0.0506 | No | ||
| 72 | PAK1IP1 | 1423766_at 1430875_a_at | 9886 | 0.033 | 0.0401 | No | ||
| 73 | TLR4 | 1418162_at 1418163_at 1430695_at 1442827_at | 9897 | 0.033 | 0.0400 | No | ||
| 74 | TCTA | 1420123_at 1420124_s_at 1420125_at 1424068_at | 10235 | 0.011 | 0.0246 | No | ||
| 75 | ACVRL1 | 1435825_at 1451604_a_at | 10243 | 0.010 | 0.0244 | No | ||
| 76 | DPP8 | 1428697_at 1431064_at 1445705_x_at | 10261 | 0.010 | 0.0237 | No | ||
| 77 | RAB20 | 1438097_at | 10263 | 0.009 | 0.0237 | No | ||
| 78 | SSTR5 | 1445459_at | 10295 | 0.007 | 0.0224 | No | ||
| 79 | SCRT2 | 1440930_a_at 1440931_at 1446986_at 1457310_x_at | 10320 | 0.006 | 0.0213 | No | ||
| 80 | ARID1A | 1447104_at | 10559 | -0.009 | 0.0105 | No | ||
| 81 | ELOVL5 | 1415840_at 1437211_x_at 1459242_at | 10880 | -0.030 | -0.0039 | No | ||
| 82 | PPP1R14C | 1417701_at 1443799_at 1446671_at | 10890 | -0.031 | -0.0040 | No | ||
| 83 | TRPM3 | 1439026_at 1441966_at 1443723_at 1445555_at 1456633_at 1456923_at 1457927_at | 11065 | -0.042 | -0.0116 | No | ||
| 84 | PTK7 | 1452589_at | 11149 | -0.047 | -0.0150 | No | ||
| 85 | MTPN | 1420472_at 1420473_at 1420474_at 1420475_at 1437457_a_at 1459597_at | 11206 | -0.050 | -0.0171 | No | ||
| 86 | SLC9A3R1 | 1438115_a_at 1438116_x_at 1450982_at | 11284 | -0.056 | -0.0200 | No | ||
| 87 | CDKN2A | 1450140_a_at | 11446 | -0.065 | -0.0268 | No | ||
| 88 | PCSK5 | 1424605_at 1437339_s_at 1438248_at 1451406_a_at 1452475_at 1459124_at | 11661 | -0.079 | -0.0359 | No | ||
| 89 | TWIST1 | 1418733_at | 11770 | -0.085 | -0.0400 | No | ||
| 90 | IRAK4 | 1421670_a_at 1451749_at | 12627 | -0.137 | -0.0780 | No | ||
| 91 | ATP5H | 1423676_at 1435112_a_at | 12685 | -0.141 | -0.0792 | No | ||
| 92 | NCF2 | 1448561_at | 12797 | -0.149 | -0.0829 | No | ||
| 93 | ZIC2 | 1421301_at | 12872 | -0.154 | -0.0848 | No | ||
| 94 | PTPRCAP | 1448511_at | 13138 | -0.170 | -0.0953 | No | ||
| 95 | NEURL2 | 1429141_at | 13240 | -0.176 | -0.0982 | No | ||
| 96 | TBXAS1 | 1416827_at | 13749 | -0.211 | -0.1194 | No | ||
| 97 | IL18 | 1417932_at | 13852 | -0.218 | -0.1220 | No | ||
| 98 | FAU | 1417452_a_at | 14044 | -0.231 | -0.1285 | No | ||
| 99 | PLCB2 | 1452481_at | 14116 | -0.236 | -0.1295 | No | ||
| 100 | GPR97 | 1417894_at | 14300 | -0.248 | -0.1355 | No | ||
| 101 | LAMA3 | 1444860_at | 14338 | -0.251 | -0.1347 | No | ||
| 102 | CREB5 | 1440089_at 1442576_at 1457222_at | 14625 | -0.273 | -0.1452 | No | ||
| 103 | TNFSF13 | 1445227_at | 14971 | -0.298 | -0.1581 | No | ||
| 104 | DGKZ | 1426738_at 1452169_a_at | 15090 | -0.306 | -0.1605 | No | ||
| 105 | CTSG | 1419594_at | 15263 | -0.319 | -0.1653 | No | ||
| 106 | LTB4R | 1420407_at | 15296 | -0.322 | -0.1637 | No | ||
| 107 | FGF12 | 1440270_at 1451693_a_at 1460137_at | 15527 | -0.338 | -0.1709 | No | ||
| 108 | MSH5 | 1430771_a_at 1449537_at | 15562 | -0.341 | -0.1691 | No | ||
| 109 | LAIR1 | 1430447_a_at 1439067_at 1444040_at | 15648 | -0.348 | -0.1696 | No | ||
| 110 | PHOX2B | 1422232_at 1455907_x_at | 15743 | -0.354 | -0.1705 | No | ||
| 111 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 15846 | -0.364 | -0.1716 | No | ||
| 112 | JARID2 | 1422698_s_at 1446082_at 1450710_at 1456661_at | 15855 | -0.365 | -0.1684 | No | ||
| 113 | RHOA | 1437628_s_at | 15960 | -0.374 | -0.1696 | No | ||
| 114 | ADAM11 | 1450248_at | 16064 | -0.382 | -0.1706 | No | ||
| 115 | MPO | 1415960_at | 16160 | -0.390 | -0.1711 | No | ||
| 116 | SESN2 | 1425139_at 1451599_at | 16245 | -0.398 | -0.1711 | No | ||
| 117 | SSBP4 | 1449511_a_at | 16306 | -0.402 | -0.1699 | No | ||
| 118 | CCT7 | 1415816_at 1415817_s_at | 16463 | -0.418 | -0.1730 | No | ||
| 119 | NFKB2 | 1425902_a_at 1429128_x_at | 16780 | -0.446 | -0.1832 | No | ||
| 120 | RERG | 1447446_at 1451236_at | 17125 | -0.481 | -0.1943 | No | ||
| 121 | EIF3S6 | 1419822_at 1434523_x_at 1439268_x_at 1460432_a_at | 17505 | -0.522 | -0.2066 | No | ||
| 122 | ARHGEF4 | 1435033_at 1437656_at | 17654 | -0.539 | -0.2081 | No | ||
| 123 | MRPS18B | 1451164_a_at | 17664 | -0.540 | -0.2032 | No | ||
| 124 | STAT6 | 1421708_a_at 1426353_at | 17837 | -0.561 | -0.2057 | No | ||
| 125 | CHRM4 | 1450575_at | 17915 | -0.572 | -0.2036 | No | ||
| 126 | H1FX | 1436517_at | 17924 | -0.574 | -0.1984 | No | ||
| 127 | ONECUT1 | 1421447_at 1450252_at 1456974_at | 18064 | -0.592 | -0.1990 | No | ||
| 128 | MRPL49 | 1423218_a_at 1423219_a_at | 18129 | -0.601 | -0.1960 | No | ||
| 129 | PBX1 | 1430183_at 1440037_at 1440954_at 1442846_at 1449542_at 1458853_at | 18265 | -0.619 | -0.1962 | No | ||
| 130 | SEPT1 | 1449898_at | 18413 | -0.638 | -0.1967 | No | ||
| 131 | VDAC2 | 1415990_at | 18484 | -0.649 | -0.1936 | No | ||
| 132 | EIF4EBP1 | 1417562_at 1417563_at 1434976_x_at | 18621 | -0.670 | -0.1933 | No | ||
| 133 | CD37 | 1419206_at 1425736_at | 18640 | -0.673 | -0.1875 | No | ||
| 134 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 18677 | -0.679 | -0.1826 | No | ||
| 135 | MAPK3 | 1427060_at | 18852 | -0.706 | -0.1836 | No | ||
| 136 | SLC9A1 | 1417397_at | 18930 | -0.718 | -0.1802 | No | ||
| 137 | GNGT2 | 1428733_at | 18993 | -0.729 | -0.1759 | No | ||
| 138 | ATP2A3 | 1421129_a_at 1450124_a_at | 19017 | -0.733 | -0.1698 | No | ||
| 139 | RNF111 | 1423304_a_at | 19022 | -0.733 | -0.1628 | No | ||
| 140 | SEMA6B | 1448790_at | 19072 | -0.742 | -0.1578 | No | ||
| 141 | CBX8 | 1419198_at 1419199_at | 19304 | -0.785 | -0.1608 | No | ||
| 142 | GATA3 | 1448886_at | 19704 | -0.862 | -0.1707 | No | ||
| 143 | ELMO1 | 1424523_at 1446610_at 1450208_a_at | 19899 | -0.907 | -0.1707 | No | ||
| 144 | USP46 | 1435325_at 1436518_at 1440416_at | 19913 | -0.911 | -0.1624 | No | ||
| 145 | SLC16A13 | 1453056_at | 19926 | -0.914 | -0.1540 | No | ||
| 146 | TNFSF11 | 1419083_at 1451944_a_at | 20051 | -0.945 | -0.1505 | No | ||
| 147 | VAV1 | 1422932_a_at | 20103 | -0.957 | -0.1435 | No | ||
| 148 | ERF | 1422114_at 1435561_at | 20175 | -0.974 | -0.1372 | No | ||
| 149 | SLC27A3 | 1427180_at | 20255 | -0.996 | -0.1311 | No | ||
| 150 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 20268 | -1.000 | -0.1219 | No | ||
| 151 | BIRC3 | 1421392_a_at 1425223_at | 20286 | -1.002 | -0.1129 | No | ||
| 152 | ABI1 | 1423177_a_at 1423178_at 1450890_a_at | 20537 | -1.058 | -0.1141 | No | ||
| 153 | KIFC3 | 1416199_at | 20552 | -1.064 | -0.1043 | No | ||
| 154 | POLD4 | 1427885_at | 20622 | -1.085 | -0.0969 | No | ||
| 155 | PPGB | 1445659_at 1448128_at 1458466_at | 20769 | -1.142 | -0.0924 | No | ||
| 156 | PCQAP | 1441617_at 1444680_at 1444688_at 1448435_at | 21155 | -1.342 | -0.0970 | No | ||
| 157 | LCP2 | 1418641_at 1418642_at | 21385 | -1.531 | -0.0926 | No | ||
| 158 | LTA | 1420353_at | 21540 | -1.710 | -0.0829 | No | ||
| 159 | ETV1 | 1422607_at 1445314_at 1450684_at | 21566 | -1.747 | -0.0670 | No | ||
| 160 | PDLIM2 | 1423946_at | 21685 | -2.019 | -0.0527 | No | ||
| 161 | SESN3 | 1449303_at 1453313_at | 21827 | -2.823 | -0.0316 | No | ||
| 162 | CKB | 1455106_a_at | 21906 | -3.736 | 0.0013 | No |