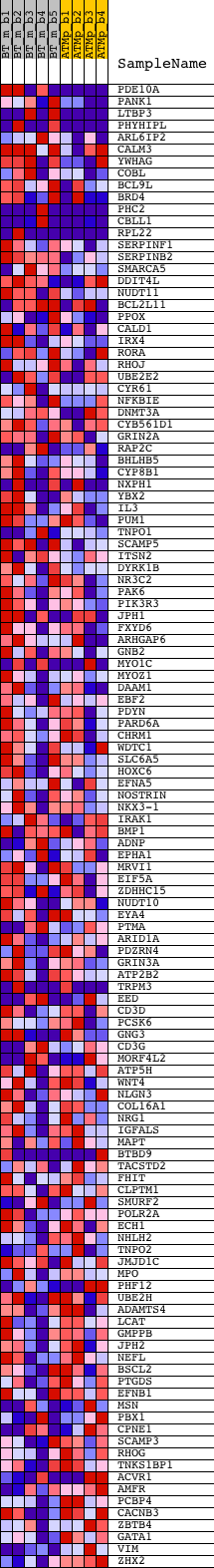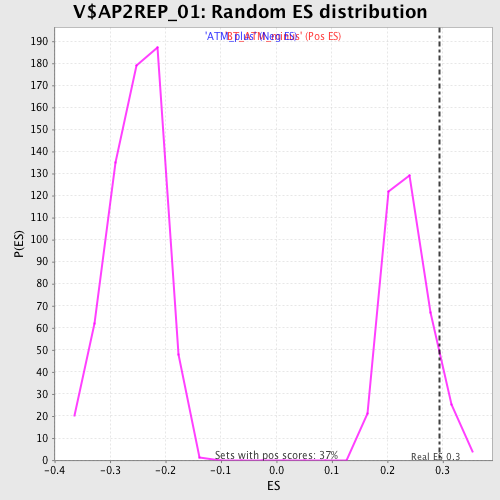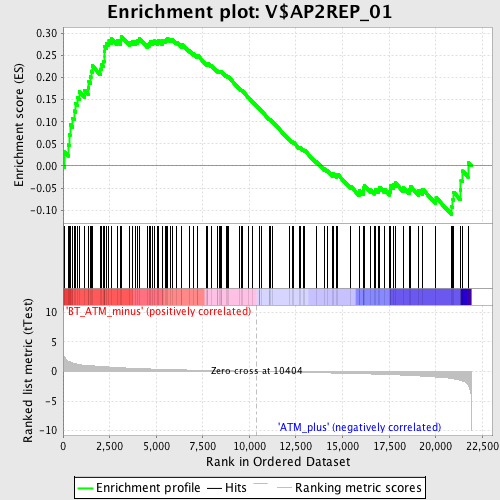
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$AP2REP_01 |
| Enrichment Score (ES) | 0.29365832 |
| Normalized Enrichment Score (NES) | 1.2493489 |
| Nominal p-value | 0.07880435 |
| FDR q-value | 0.5256228 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PDE10A | 1419389_at 1419390_at 1432490_a_at 1439618_at 1458499_at | 90 | 2.370 | 0.0317 | Yes | ||
| 2 | PANK1 | 1418715_at 1429813_at 1429814_at 1431028_a_at 1457110_at | 297 | 1.676 | 0.0476 | Yes | ||
| 3 | LTBP3 | 1418049_at 1437833_at 1438312_s_at 1456189_x_at | 323 | 1.649 | 0.0714 | Yes | ||
| 4 | PHYHIPL | 1427023_at 1437062_s_at | 374 | 1.606 | 0.0933 | Yes | ||
| 5 | ARL6IP2 | 1416793_at 1416794_at 1431197_at 1435594_at | 519 | 1.424 | 0.1083 | Yes | ||
| 6 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 608 | 1.338 | 0.1244 | Yes | ||
| 7 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 675 | 1.285 | 0.1408 | Yes | ||
| 8 | COBL | 1434917_at 1442076_at | 767 | 1.230 | 0.1552 | Yes | ||
| 9 | BCL9L | 1419180_at 1438806_at 1443700_at 1444855_at | 874 | 1.157 | 0.1679 | Yes | ||
| 10 | BRD4 | 1424922_a_at 1440078_at 1450711_at 1451870_a_at 1458708_at | 1143 | 1.046 | 0.1714 | Yes | ||
| 11 | PHC2 | 1416048_at 1437239_x_at 1439639_at | 1345 | 1.000 | 0.1773 | Yes | ||
| 12 | CBLL1 | 1421122_at 1437203_at 1458261_at | 1383 | 0.994 | 0.1906 | Yes | ||
| 13 | RPL22 | 1448398_s_at 1453118_s_at | 1460 | 0.978 | 0.2019 | Yes | ||
| 14 | SERPINF1 | 1416168_at 1453724_a_at | 1500 | 0.969 | 0.2148 | Yes | ||
| 15 | SERPINB2 | 1419082_at | 1558 | 0.957 | 0.2266 | Yes | ||
| 16 | SMARCA5 | 1440048_at | 2000 | 0.855 | 0.2193 | Yes | ||
| 17 | DDIT4L | 1439332_at 1444139_at 1451751_at | 2044 | 0.846 | 0.2301 | Yes | ||
| 18 | NUDT11 | 1426887_at | 2161 | 0.818 | 0.2372 | Yes | ||
| 19 | BCL2L11 | 1426334_a_at 1435448_at 1435449_at 1456005_a_at 1456006_at 1459794_at | 2208 | 0.806 | 0.2472 | Yes | ||
| 20 | PPOX | 1416618_at 1441862_at 1448065_at | 2210 | 0.806 | 0.2594 | Yes | ||
| 21 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 2233 | 0.801 | 0.2705 | Yes | ||
| 22 | IRX4 | 1419539_at | 2327 | 0.781 | 0.2780 | Yes | ||
| 23 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 2462 | 0.751 | 0.2832 | Yes | ||
| 24 | RHOJ | 1418892_at 1444982_at | 2586 | 0.723 | 0.2885 | Yes | ||
| 25 | UBE2E2 | 1424358_at 1447025_at | 2898 | 0.665 | 0.2843 | Yes | ||
| 26 | CYR61 | 1416039_x_at 1417848_at 1417849_at 1438133_a_at 1442340_x_at 1457823_at 1459462_at | 3095 | 0.628 | 0.2848 | Yes | ||
| 27 | NFKBIE | 1431843_a_at 1458299_s_at | 3109 | 0.625 | 0.2937 | Yes | ||
| 28 | DNMT3A | 1423063_at 1423064_at 1423065_at 1423066_at 1442309_at 1460324_at | 3583 | 0.552 | 0.2803 | No | ||
| 29 | CYB561D1 | 1431021_at 1434226_at 1459837_at | 3730 | 0.531 | 0.2816 | No | ||
| 30 | GRIN2A | 1421616_at | 3884 | 0.512 | 0.2824 | No | ||
| 31 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 4011 | 0.498 | 0.2841 | No | ||
| 32 | BHLHB5 | 1418271_at | 4092 | 0.486 | 0.2878 | No | ||
| 33 | CYP8B1 | 1449309_at | 4539 | 0.436 | 0.2739 | No | ||
| 34 | NXPH1 | 1436718_at 1438388_at 1459921_at | 4617 | 0.428 | 0.2769 | No | ||
| 35 | YBX2 | 1420762_a_at | 4670 | 0.424 | 0.2809 | No | ||
| 36 | IL3 | 1450566_at | 4802 | 0.410 | 0.2811 | No | ||
| 37 | PUM1 | 1423117_at 1456054_a_at 1460325_at | 4896 | 0.401 | 0.2829 | No | ||
| 38 | TNPO1 | 1433585_at 1447034_at 1455043_at | 5093 | 0.381 | 0.2796 | No | ||
| 39 | SCAMP5 | 1450247_a_at 1451224_at | 5128 | 0.378 | 0.2838 | No | ||
| 40 | ITSN2 | 1423184_at 1431772_a_at 1435023_at 1445375_at 1446595_at 1446735_at 1446911_at | 5344 | 0.358 | 0.2794 | No | ||
| 41 | DYRK1B | 1422254_a_at | 5352 | 0.358 | 0.2844 | No | ||
| 42 | NR3C2 | 1435991_at 1440306_at 1442533_at | 5479 | 0.346 | 0.2839 | No | ||
| 43 | PAK6 | 1445475_at 1455200_at | 5533 | 0.342 | 0.2866 | No | ||
| 44 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 5592 | 0.337 | 0.2891 | No | ||
| 45 | JPH1 | 1421520_at 1438428_at | 5765 | 0.321 | 0.2860 | No | ||
| 46 | FXYD6 | 1417343_at | 5856 | 0.311 | 0.2866 | No | ||
| 47 | ARHGAP6 | 1417704_a_at 1451867_x_at 1456333_a_at | 6094 | 0.293 | 0.2802 | No | ||
| 48 | GNB2 | 1450623_at | 6377 | 0.271 | 0.2713 | No | ||
| 49 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 6378 | 0.271 | 0.2754 | No | ||
| 50 | MYOZ1 | 1448636_at 1460202_at | 6770 | 0.240 | 0.2611 | No | ||
| 51 | DAAM1 | 1431035_at 1455244_at 1458662_at | 7004 | 0.222 | 0.2538 | No | ||
| 52 | EBF2 | 1418494_at 1446128_at 1449101_at 1449102_at | 7190 | 0.209 | 0.2485 | No | ||
| 53 | PDYN | 1416266_at | 7214 | 0.207 | 0.2506 | No | ||
| 54 | PARD6A | 1449100_at | 7708 | 0.173 | 0.2306 | No | ||
| 55 | CHRM1 | 1439611_at 1450833_at | 7765 | 0.170 | 0.2306 | No | ||
| 56 | WDTC1 | 1434560_at | 7775 | 0.170 | 0.2327 | No | ||
| 57 | SLC6A5 | 1425992_at | 7951 | 0.158 | 0.2271 | No | ||
| 58 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 8275 | 0.134 | 0.2143 | No | ||
| 59 | EFNA5 | 1421796_a_at 1436866_at 1440052_at 1446383_at 1451930_at | 8307 | 0.133 | 0.2149 | No | ||
| 60 | NOSTRIN | 1441075_at | 8373 | 0.128 | 0.2139 | No | ||
| 61 | NKX3-1 | 1449998_at | 8437 | 0.124 | 0.2129 | No | ||
| 62 | IRAK1 | 1448668_a_at 1460649_at | 8492 | 0.121 | 0.2122 | No | ||
| 63 | BMP1 | 1426238_at 1427457_a_at 1427458_at 1446007_at | 8513 | 0.120 | 0.2131 | No | ||
| 64 | ADNP | 1423069_at | 8747 | 0.104 | 0.2040 | No | ||
| 65 | EPHA1 | 1422917_at 1432399_a_at 1432400_at | 8802 | 0.101 | 0.2030 | No | ||
| 66 | MRVI1 | 1422245_a_at 1459665_s_at | 8868 | 0.098 | 0.2015 | No | ||
| 67 | EIF5A | 1437859_x_at 1451470_s_at | 9451 | 0.061 | 0.1758 | No | ||
| 68 | ZDHHC15 | 1442274_at | 9562 | 0.054 | 0.1716 | No | ||
| 69 | NUDT10 | 1435061_at | 9611 | 0.051 | 0.1701 | No | ||
| 70 | EYA4 | 1432461_at 1442542_at 1445809_at 1456647_a_at 1456665_at | 9960 | 0.029 | 0.1546 | No | ||
| 71 | PTMA | 1423455_at | 10172 | 0.014 | 0.1452 | No | ||
| 72 | ARID1A | 1447104_at | 10559 | -0.009 | 0.1276 | No | ||
| 73 | PDZRN4 | 1456512_at | 10560 | -0.009 | 0.1277 | No | ||
| 74 | GRIN3A | 1436575_at 1438866_at 1458378_at | 10563 | -0.009 | 0.1278 | No | ||
| 75 | ATP2B2 | 1420402_at 1420403_at 1433888_at | 10629 | -0.014 | 0.1250 | No | ||
| 76 | TRPM3 | 1439026_at 1441966_at 1443723_at 1445555_at 1456633_at 1456923_at 1457927_at | 11065 | -0.042 | 0.1057 | No | ||
| 77 | EED | 1448653_at | 11091 | -0.044 | 0.1052 | No | ||
| 78 | CD3D | 1422828_at | 11144 | -0.046 | 0.1035 | No | ||
| 79 | PCSK6 | 1426981_at | 11254 | -0.054 | 0.0993 | No | ||
| 80 | GNG3 | 1417428_at | 12143 | -0.107 | 0.0602 | No | ||
| 81 | CD3G | 1419178_at | 12305 | -0.117 | 0.0546 | No | ||
| 82 | MORF4L2 | 1415778_at 1439413_x_at | 12379 | -0.123 | 0.0531 | No | ||
| 83 | ATP5H | 1423676_at 1435112_a_at | 12685 | -0.141 | 0.0413 | No | ||
| 84 | WNT4 | 1450782_at | 12722 | -0.144 | 0.0418 | No | ||
| 85 | NLGN3 | 1456384_at | 12897 | -0.155 | 0.0362 | No | ||
| 86 | COL16A1 | 1427986_a_at | 12972 | -0.159 | 0.0352 | No | ||
| 87 | NRG1 | 1456524_at | 13601 | -0.201 | 0.0094 | No | ||
| 88 | IGFALS | 1422826_at | 14024 | -0.229 | -0.0064 | No | ||
| 89 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 14182 | -0.240 | -0.0100 | No | ||
| 90 | BTBD9 | 1431230_a_at 1434769_at 1435706_at 1440548_at 1444827_at 1445220_at 1458043_at | 14470 | -0.261 | -0.0192 | No | ||
| 91 | TACSTD2 | 1423323_at | 14522 | -0.265 | -0.0176 | No | ||
| 92 | FHIT | 1425893_a_at | 14705 | -0.279 | -0.0217 | No | ||
| 93 | CLPTM1 | 1416883_at | 14748 | -0.282 | -0.0194 | No | ||
| 94 | SMURF2 | 1429045_at 1429046_at 1454894_at | 15458 | -0.334 | -0.0468 | No | ||
| 95 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 15897 | -0.369 | -0.0613 | No | ||
| 96 | ECH1 | 1448491_at | 15904 | -0.370 | -0.0560 | No | ||
| 97 | NHLH2 | 1436888_at | 16115 | -0.386 | -0.0598 | No | ||
| 98 | TNPO2 | 1425592_at 1425593_at 1437283_at | 16119 | -0.386 | -0.0541 | No | ||
| 99 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 16149 | -0.389 | -0.0496 | No | ||
| 100 | MPO | 1415960_at | 16160 | -0.390 | -0.0441 | No | ||
| 101 | PHF12 | 1434922_at 1453579_at | 16518 | -0.422 | -0.0541 | No | ||
| 102 | UBE2H | 1418631_at 1418632_at 1428791_at 1438971_x_at 1447374_at 1459930_at | 16727 | -0.440 | -0.0570 | No | ||
| 103 | ADAMTS4 | 1452595_at | 16792 | -0.447 | -0.0532 | No | ||
| 104 | LCAT | 1417043_at | 16960 | -0.464 | -0.0538 | No | ||
| 105 | GMPPB | 1439030_at | 17005 | -0.469 | -0.0488 | No | ||
| 106 | JPH2 | 1421453_at 1455404_at | 17244 | -0.494 | -0.0522 | No | ||
| 107 | NEFL | 1426255_at | 17545 | -0.526 | -0.0580 | No | ||
| 108 | BSCL2 | 1420632_a_at 1437345_a_at | 17578 | -0.529 | -0.0515 | No | ||
| 109 | PTGDS | 1423859_a_at 1423860_at | 17602 | -0.532 | -0.0445 | No | ||
| 110 | EFNB1 | 1418285_at 1418286_a_at 1451591_a_at | 17742 | -0.549 | -0.0426 | No | ||
| 111 | MSN | 1421814_at 1450379_at | 17833 | -0.561 | -0.0382 | No | ||
| 112 | PBX1 | 1430183_at 1440037_at 1440954_at 1442846_at 1449542_at 1458853_at | 18265 | -0.619 | -0.0486 | No | ||
| 113 | CPNE1 | 1433439_at | 18590 | -0.666 | -0.0534 | No | ||
| 114 | SCAMP3 | 1416294_at | 18647 | -0.675 | -0.0458 | No | ||
| 115 | RHOG | 1422572_at | 19100 | -0.748 | -0.0552 | No | ||
| 116 | TNKS1BP1 | 1427051_at | 19294 | -0.783 | -0.0522 | No | ||
| 117 | ACVR1 | 1416786_at 1416787_at 1448460_at 1457551_at | 20023 | -0.938 | -0.0714 | No | ||
| 118 | AMFR | 1450102_a_at | 20862 | -1.184 | -0.0920 | No | ||
| 119 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20933 | -1.222 | -0.0767 | No | ||
| 120 | CACNB3 | 1448656_at | 20963 | -1.239 | -0.0593 | No | ||
| 121 | ZBTB4 | 1429722_at 1460488_at | 21344 | -1.502 | -0.0540 | No | ||
| 122 | GATA1 | 1449232_at | 21364 | -1.516 | -0.0320 | No | ||
| 123 | VIM | 1438118_x_at 1450641_at 1456292_a_at | 21441 | -1.597 | -0.0114 | No | ||
| 124 | ZHX2 | 1455210_at | 21747 | -2.244 | 0.0086 | No |