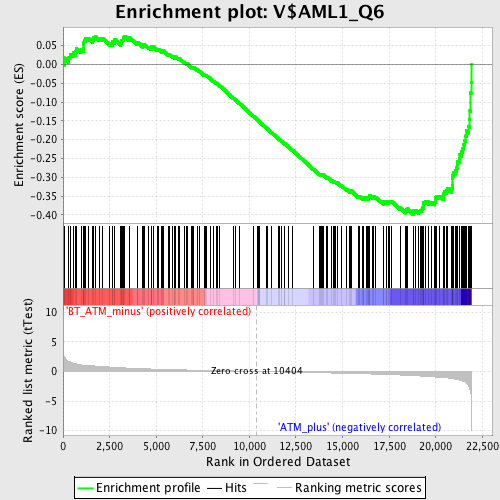
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$AML1_Q6 |
| Enrichment Score (ES) | -0.39868718 |
| Normalized Enrichment Score (NES) | -1.6697302 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.039094035 |
| FWER p-Value | 0.383 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 50 | 2.528 | 0.0170 | No | ||
| 2 | PTK9 | 1420873_at 1420874_at 1420875_at | 310 | 1.662 | 0.0178 | No | ||
| 3 | MYOD1 | 1418420_at 1447675_x_at | 372 | 1.607 | 0.0272 | No | ||
| 4 | COL4A1 | 1426348_at 1452035_at | 535 | 1.408 | 0.0305 | No | ||
| 5 | GPATC2 | 1420528_at 1440329_s_at 1445189_at 1452498_at 1458566_at | 679 | 1.284 | 0.0337 | No | ||
| 6 | JDP2 | 1424972_at 1450350_a_at | 708 | 1.267 | 0.0421 | No | ||
| 7 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 961 | 1.114 | 0.0390 | No | ||
| 8 | SLC12A6 | 1436989_s_at 1449878_a_at 1458916_at | 1091 | 1.063 | 0.0412 | No | ||
| 9 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 1100 | 1.059 | 0.0489 | No | ||
| 10 | GZMB | 1419060_at | 1103 | 1.058 | 0.0569 | No | ||
| 11 | DIABLO | 1417683_at 1425768_at | 1121 | 1.052 | 0.0641 | No | ||
| 12 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 1207 | 1.030 | 0.0681 | No | ||
| 13 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 1340 | 1.001 | 0.0697 | No | ||
| 14 | RORC | 1425792_a_at 1425793_a_at | 1595 | 0.950 | 0.0652 | No | ||
| 15 | CTSK | 1450652_at | 1639 | 0.939 | 0.0704 | No | ||
| 16 | ABCD4 | 1419572_a_at 1427829_at | 1728 | 0.919 | 0.0734 | No | ||
| 17 | CORO1C | 1417752_at 1419911_at 1437721_at 1442972_at 1449660_s_at 1459169_at | 1968 | 0.864 | 0.0690 | No | ||
| 18 | SLC37A2 | 1452492_a_at | 2106 | 0.832 | 0.0690 | No | ||
| 19 | CHES1 | 1434002_at 1436925_at 1438255_at 1440607_at 1444555_at 1447325_at 1453094_at | 2506 | 0.741 | 0.0563 | No | ||
| 20 | KCNJ1 | 1418613_at 1418614_at | 2632 | 0.714 | 0.0561 | No | ||
| 21 | PTPN22 | 1417995_at 1442820_at | 2637 | 0.713 | 0.0613 | No | ||
| 22 | SCEL | 1422837_at 1459426_at | 2771 | 0.686 | 0.0604 | No | ||
| 23 | GTF2A1 | 1421357_at | 2776 | 0.686 | 0.0655 | No | ||
| 24 | SUPT16H | 1419741_at 1449578_at 1456449_at | 3107 | 0.625 | 0.0551 | No | ||
| 25 | LMO2 | 1454086_a_at | 3128 | 0.623 | 0.0589 | No | ||
| 26 | ARHGAP21 | 1428368_at 1428369_s_at 1445628_at 1447206_at | 3136 | 0.622 | 0.0633 | No | ||
| 27 | MMP14 | 1416572_at 1448383_at | 3213 | 0.608 | 0.0645 | No | ||
| 28 | CYP17A1 | 1417017_at | 3219 | 0.607 | 0.0689 | No | ||
| 29 | COL9A2 | 1450673_at | 3246 | 0.601 | 0.0723 | No | ||
| 30 | MR1 | 1421898_a_at 1421899_a_at | 3288 | 0.597 | 0.0749 | No | ||
| 31 | ARHGAP12 | 1431052_at 1444463_at 1451525_at 1451526_at | 3558 | 0.555 | 0.0668 | No | ||
| 32 | RAB5B | 1422119_at 1437446_at | 3576 | 0.553 | 0.0702 | No | ||
| 33 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 3986 | 0.501 | 0.0553 | No | ||
| 34 | SCNN1A | 1425088_at | 4018 | 0.498 | 0.0576 | No | ||
| 35 | ARNT | 1419996_s_at 1421721_a_at 1425064_at 1437042_at 1449696_at | 4280 | 0.465 | 0.0492 | No | ||
| 36 | SCN2B | 1430648_at 1436134_at 1457144_at | 4301 | 0.461 | 0.0518 | No | ||
| 37 | SCN3B | 1426328_a_at 1435767_at | 4379 | 0.454 | 0.0517 | No | ||
| 38 | NUCB2 | 1418355_at | 4610 | 0.429 | 0.0444 | No | ||
| 39 | MTX1 | 1418521_a_at 1418522_at 1447361_at | 4734 | 0.417 | 0.0419 | No | ||
| 40 | SIRT1 | 1418640_at 1458538_at | 4742 | 0.416 | 0.0448 | No | ||
| 41 | ITGA10 | 1440235_at | 4762 | 0.414 | 0.0471 | No | ||
| 42 | EMP1 | 1416529_at 1459171_at | 4844 | 0.405 | 0.0464 | No | ||
| 43 | UBL3 | 1423461_a_at | 5068 | 0.384 | 0.0391 | No | ||
| 44 | PICALM | 1441328_at 1443218_at 1446968_at 1451316_a_at 1455773_at | 5120 | 0.379 | 0.0397 | No | ||
| 45 | RIN1 | 1424507_at 1437460_x_at | 5287 | 0.364 | 0.0348 | No | ||
| 46 | HOXC4 | 1422870_at | 5361 | 0.356 | 0.0342 | No | ||
| 47 | TLL2 | 1421526_at | 5365 | 0.356 | 0.0368 | No | ||
| 48 | NGB | 1417996_at 1417997_at | 5682 | 0.328 | 0.0247 | No | ||
| 49 | ANK3 | 1425202_a_at 1439220_at 1447259_at 1451628_a_at 1457288_at | 5718 | 0.325 | 0.0256 | No | ||
| 50 | ZBTB8 | 1455193_at | 5863 | 0.311 | 0.0214 | No | ||
| 51 | AKAP3 | 1421531_at 1456591_x_at | 5999 | 0.300 | 0.0174 | No | ||
| 52 | BLOC1S2 | 1429048_at | 6003 | 0.300 | 0.0196 | No | ||
| 53 | LUZP1 | 1416469_at 1448352_at | 6023 | 0.298 | 0.0210 | No | ||
| 54 | SLC2A3 | 1421924_at 1427770_a_at 1437052_s_at 1455898_x_at | 6205 | 0.284 | 0.0148 | No | ||
| 55 | SCN8A | 1423515_at 1439889_at 1442208_at 1451894_a_at 1457412_at 1458693_at | 6238 | 0.281 | 0.0155 | No | ||
| 56 | RGS1 | 1417601_at | 6520 | 0.259 | 0.0046 | No | ||
| 57 | MAMDC1 | 1442206_at 1442561_at 1445372_at 1458147_at | 6626 | 0.251 | 0.0017 | No | ||
| 58 | CREM | 1418322_at 1430598_at 1430847_a_at 1449037_at | 6668 | 0.248 | 0.0017 | No | ||
| 59 | PDZK1 | 1431701_a_at | 6918 | 0.229 | -0.0080 | No | ||
| 60 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 6943 | 0.227 | -0.0074 | No | ||
| 61 | SLITRK5 | 1429718_at | 7019 | 0.221 | -0.0092 | No | ||
| 62 | FCER1G | 1418340_at | 7020 | 0.221 | -0.0075 | No | ||
| 63 | RCOR2 | 1417302_at | 7191 | 0.209 | -0.0137 | No | ||
| 64 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 7337 | 0.200 | -0.0188 | No | ||
| 65 | CUGBP1 | 1423932_at 1425932_a_at 1426407_at 1426408_at 1427413_a_at | 7584 | 0.182 | -0.0288 | No | ||
| 66 | KCTD4 | 1420537_at 1441801_at | 7598 | 0.182 | -0.0280 | No | ||
| 67 | RICS | 1438451_at 1447099_at | 7654 | 0.177 | -0.0291 | No | ||
| 68 | DDX23 | 1430050_at | 7679 | 0.175 | -0.0289 | No | ||
| 69 | NRAS | 1422688_a_at 1454060_a_at | 7939 | 0.159 | -0.0396 | No | ||
| 70 | PDE3B | 1433694_at | 8088 | 0.147 | -0.0453 | No | ||
| 71 | FOXD2 | 1450289_at | 8232 | 0.137 | -0.0508 | No | ||
| 72 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 8275 | 0.134 | -0.0517 | No | ||
| 73 | IL23A | 1419529_at | 8411 | 0.126 | -0.0570 | No | ||
| 74 | DAB1 | 1421100_a_at 1427307_a_at 1427308_at 1435577_at 1435578_s_at | 9159 | 0.080 | -0.0907 | No | ||
| 75 | THBS3 | 1416623_at | 9234 | 0.075 | -0.0935 | No | ||
| 76 | PRICKLE1 | 1441663_at 1442172_at 1442400_at 1444759_at 1452249_at 1457637_at | 9454 | 0.061 | -0.1031 | No | ||
| 77 | IFNG | 1425947_at | 10234 | 0.011 | -0.1389 | No | ||
| 78 | NAV3 | 1459440_at | 10451 | -0.003 | -0.1488 | No | ||
| 79 | MMP13 | 1417256_at | 10497 | -0.005 | -0.1508 | No | ||
| 80 | AGTRL1 | 1423037_at 1438651_a_at | 10511 | -0.006 | -0.1514 | No | ||
| 81 | PDZRN4 | 1456512_at | 10560 | -0.009 | -0.1535 | No | ||
| 82 | DKK1 | 1420360_at 1458232_at | 10571 | -0.010 | -0.1539 | No | ||
| 83 | CLDN10 | 1426147_s_at | 10915 | -0.033 | -0.1694 | No | ||
| 84 | MIZF | 1440255_at | 10995 | -0.038 | -0.1727 | No | ||
| 85 | FGD4 | 1425037_at 1426041_a_at 1426042_at 1451659_at 1455337_at | 11188 | -0.049 | -0.1812 | No | ||
| 86 | MPL | 1421461_at 1451919_a_at | 11548 | -0.071 | -0.1972 | No | ||
| 87 | MOAP1 | 1431823_at 1448787_at | 11609 | -0.075 | -0.1993 | No | ||
| 88 | HDGF | 1415888_at 1419964_s_at 1449681_at | 11732 | -0.083 | -0.2043 | No | ||
| 89 | MATN1 | 1418477_at | 11902 | -0.093 | -0.2114 | No | ||
| 90 | DNM3 | 1425403_at 1436624_at 1436875_at 1438801_at 1445540_at 1446265_at 1446431_at 1446694_at 1457405_at | 11909 | -0.093 | -0.2109 | No | ||
| 91 | TBX5 | 1425694_at 1425695_at | 12080 | -0.103 | -0.2180 | No | ||
| 92 | WNT8B | 1421439_at | 12331 | -0.119 | -0.2285 | No | ||
| 93 | MAP3K11 | 1450669_at | 13441 | -0.190 | -0.2781 | No | ||
| 94 | SLC6A9 | 1417636_at 1431812_a_at | 13787 | -0.213 | -0.2923 | No | ||
| 95 | RASAL2 | 1436910_at 1444671_at | 13821 | -0.216 | -0.2922 | No | ||
| 96 | EIF2B3 | 1434524_at | 13884 | -0.221 | -0.2933 | No | ||
| 97 | CRTAC1 | 1426606_at 1452936_at 1457474_at 1459858_x_at | 13930 | -0.224 | -0.2937 | No | ||
| 98 | BLOC1S1 | 1422614_s_at | 13959 | -0.225 | -0.2933 | No | ||
| 99 | NHLRC2 | 1429145_at 1433929_at 1444863_at 1453048_at 1457312_at | 14165 | -0.239 | -0.3009 | No | ||
| 100 | PGF | 1418471_at | 14171 | -0.239 | -0.2993 | No | ||
| 101 | COL4A2 | 1424051_at | 14413 | -0.256 | -0.3084 | No | ||
| 102 | TACSTD2 | 1423323_at | 14522 | -0.265 | -0.3114 | No | ||
| 103 | ANXA8 | 1417732_at 1425789_s_at | 14578 | -0.270 | -0.3118 | No | ||
| 104 | NDUFA9 | 1416663_at | 14641 | -0.274 | -0.3126 | No | ||
| 105 | KIF1B | 1423994_at 1423995_at 1425270_at 1451200_at 1451642_at 1451762_a_at 1455182_at 1458426_at | 14707 | -0.279 | -0.3134 | No | ||
| 106 | HIVEP3 | 1421150_at 1429134_at 1439660_at 1450132_at 1458802_at | 14960 | -0.297 | -0.3228 | No | ||
| 107 | GPD1 | 1416204_at 1439396_x_at 1448249_at 1456732_at | 15221 | -0.316 | -0.3323 | No | ||
| 108 | RAB39 | 1437762_at | 15375 | -0.327 | -0.3368 | No | ||
| 109 | SFRS4 | 1448778_at | 15437 | -0.332 | -0.3371 | No | ||
| 110 | EIF3S1 | 1426394_at 1426395_s_at 1452052_s_at | 15468 | -0.335 | -0.3359 | No | ||
| 111 | STRC | 1421695_at | 15847 | -0.364 | -0.3505 | No | ||
| 112 | PXN | 1424027_at 1426085_a_at 1456135_s_at | 15926 | -0.371 | -0.3513 | No | ||
| 113 | TBK1 | 1422469_at 1445571_at 1460315_s_at | 16092 | -0.385 | -0.3559 | No | ||
| 114 | MKRN3 | 1418160_at | 16133 | -0.388 | -0.3548 | No | ||
| 115 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 16149 | -0.389 | -0.3526 | No | ||
| 116 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 16310 | -0.403 | -0.3568 | No | ||
| 117 | GRASP | 1441894_s_at 1442616_at 1460206_at | 16326 | -0.404 | -0.3544 | No | ||
| 118 | ACSL5 | 1428082_at | 16421 | -0.413 | -0.3556 | No | ||
| 119 | TNFSF4 | 1421744_at | 16423 | -0.413 | -0.3525 | No | ||
| 120 | INPPL1 | 1460394_a_at | 16440 | -0.415 | -0.3501 | No | ||
| 121 | ANKRD1 | 1420991_at 1420992_at | 16447 | -0.416 | -0.3472 | No | ||
| 122 | EDARADD | 1421213_at 1433132_at 1433133_at 1437800_at | 16628 | -0.431 | -0.3522 | No | ||
| 123 | SLC37A4 | 1417042_at | 16679 | -0.436 | -0.3511 | No | ||
| 124 | ADAMTS4 | 1452595_at | 16792 | -0.447 | -0.3529 | No | ||
| 125 | BTG4 | 1426520_at | 17191 | -0.488 | -0.3674 | No | ||
| 126 | LMO3 | 1455754_at | 17198 | -0.489 | -0.3640 | No | ||
| 127 | RDH5 | 1418808_at 1421702_at | 17343 | -0.504 | -0.3668 | No | ||
| 128 | VRK1 | 1425006_a_at 1458951_at | 17389 | -0.510 | -0.3649 | No | ||
| 129 | ADAMTS8 | 1418270_at 1444174_at | 17477 | -0.520 | -0.3650 | No | ||
| 130 | ENTPD1 | 1423326_at 1450939_at 1453586_at | 17531 | -0.524 | -0.3634 | No | ||
| 131 | NRXN3 | 1419825_at 1431153_at 1432931_at 1433788_at 1438193_at 1439629_at 1442423_at 1444700_at 1456137_at 1460101_at | 17615 | -0.534 | -0.3632 | No | ||
| 132 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 18107 | -0.598 | -0.3812 | No | ||
| 133 | PDE6H | 1450765_a_at 1450766_at | 18406 | -0.637 | -0.3900 | No | ||
| 134 | RASGEF1A | 1429316_at 1435218_at | 18432 | -0.642 | -0.3863 | No | ||
| 135 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 18481 | -0.649 | -0.3835 | No | ||
| 136 | BCAR3 | 1415936_at 1431564_at | 18812 | -0.699 | -0.3934 | Yes | ||
| 137 | AP1G2 | 1419113_at | 18828 | -0.702 | -0.3887 | Yes | ||
| 138 | NFRKB | 1434753_at 1445551_at | 18921 | -0.717 | -0.3874 | Yes | ||
| 139 | RHOG | 1422572_at | 19100 | -0.748 | -0.3899 | Yes | ||
| 140 | PLXNC1 | 1423213_at 1423214_at 1443433_at 1443434_s_at 1450905_at 1450906_at | 19166 | -0.757 | -0.3871 | Yes | ||
| 141 | DGKA | 1418578_at | 19270 | -0.779 | -0.3859 | Yes | ||
| 142 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 19284 | -0.782 | -0.3806 | Yes | ||
| 143 | ATP6V0B | 1416769_s_at 1425448_x_at 1437013_x_at | 19351 | -0.793 | -0.3775 | Yes | ||
| 144 | LY9 | 1449156_at | 19355 | -0.793 | -0.3716 | Yes | ||
| 145 | TPM1 | 1423049_a_at 1423721_at 1447713_at 1456623_at | 19366 | -0.796 | -0.3660 | Yes | ||
| 146 | XCL1 | 1419412_at | 19454 | -0.812 | -0.3638 | Yes | ||
| 147 | RAB30 | 1426452_a_at | 19627 | -0.847 | -0.3653 | Yes | ||
| 148 | SNX1 | 1416260_a_at | 19787 | -0.883 | -0.3658 | Yes | ||
| 149 | RGS18 | 1420398_at 1449856_at | 19955 | -0.923 | -0.3665 | Yes | ||
| 150 | NOTCH2 | 1451889_at 1455556_at | 19986 | -0.930 | -0.3608 | Yes | ||
| 151 | DACT1 | 1417937_at | 19988 | -0.930 | -0.3537 | Yes | ||
| 152 | ARF3 | 1421789_s_at 1423973_a_at 1434787_at 1437331_a_at 1456007_at | 20058 | -0.947 | -0.3497 | Yes | ||
| 153 | IBRDC3 | 1432478_a_at 1435226_at | 20225 | -0.991 | -0.3497 | Yes | ||
| 154 | MRPS31 | 1417737_at 1443165_at | 20424 | -1.030 | -0.3510 | Yes | ||
| 155 | TITF1 | 1422346_at | 20446 | -1.035 | -0.3441 | Yes | ||
| 156 | GRK5 | 1446361_at 1446998_at 1449514_at | 20463 | -1.039 | -0.3369 | Yes | ||
| 157 | RAG1 | 1440574_at 1450680_at | 20591 | -1.074 | -0.3345 | Yes | ||
| 158 | EIF2C1 | 1434331_at | 20648 | -1.092 | -0.3288 | Yes | ||
| 159 | CD69 | 1428735_at | 20877 | -1.192 | -0.3301 | Yes | ||
| 160 | SEMA4A | 1438934_x_at 1448110_at | 20900 | -1.208 | -0.3219 | Yes | ||
| 161 | HLX1 | 1418939_at | 20908 | -1.211 | -0.3130 | Yes | ||
| 162 | SLC15A3 | 1420697_at | 20909 | -1.212 | -0.3038 | Yes | ||
| 163 | CRY1 | 1433733_a_at | 20916 | -1.215 | -0.2948 | Yes | ||
| 164 | FGF4 | 1420085_at 1420086_x_at 1449729_at 1450282_at | 20981 | -1.245 | -0.2882 | Yes | ||
| 165 | CAP1 | 1417461_at 1417462_at | 21047 | -1.281 | -0.2815 | Yes | ||
| 166 | CD6 | 1451910_a_at | 21125 | -1.321 | -0.2749 | Yes | ||
| 167 | FLI1 | 1422024_at 1433512_at 1441584_at | 21162 | -1.346 | -0.2663 | Yes | ||
| 168 | STAT2 | 1421911_at 1450403_at | 21181 | -1.359 | -0.2568 | Yes | ||
| 169 | ID3 | 1416630_at | 21267 | -1.433 | -0.2497 | Yes | ||
| 170 | MADD | 1425735_at 1455502_at | 21306 | -1.464 | -0.2403 | Yes | ||
| 171 | ACIN1 | 1416568_a_at 1421197_a_at | 21392 | -1.549 | -0.2324 | Yes | ||
| 172 | VIM | 1438118_x_at 1450641_at 1456292_a_at | 21441 | -1.597 | -0.2224 | Yes | ||
| 173 | DNAJC14 | 1426553_at 1437546_at 1437547_s_at | 21502 | -1.664 | -0.2125 | Yes | ||
| 174 | BMF | 1422995_at 1454880_s_at 1458325_x_at | 21575 | -1.757 | -0.2024 | Yes | ||
| 175 | HIPK1 | 1421298_a_at 1424540_at 1457515_at | 21619 | -1.856 | -0.1902 | Yes | ||
| 176 | BATF | 1419410_at | 21644 | -1.915 | -0.1767 | Yes | ||
| 177 | S100A9 | 1448756_at | 21769 | -2.327 | -0.1647 | Yes | ||
| 178 | TACC2 | 1425745_a_at 1447891_at | 21836 | -2.894 | -0.1456 | Yes | ||
| 179 | PACS1 | 1420112_at 1435762_at 1440014_at 1449737_at 1459707_at | 21847 | -2.951 | -0.1236 | Yes | ||
| 180 | TNNT2 | 1418726_a_at 1424967_x_at 1440424_at | 21869 | -3.198 | -0.1001 | Yes | ||
| 181 | ITGB7 | 1418741_at | 21870 | -3.202 | -0.0757 | Yes | ||
| 182 | NR4A1 | 1416505_at | 21917 | -3.996 | -0.0473 | Yes | ||
| 183 | EGR2 | 1427682_a_at 1427683_at | 21930 | -6.298 | 0.0002 | Yes |