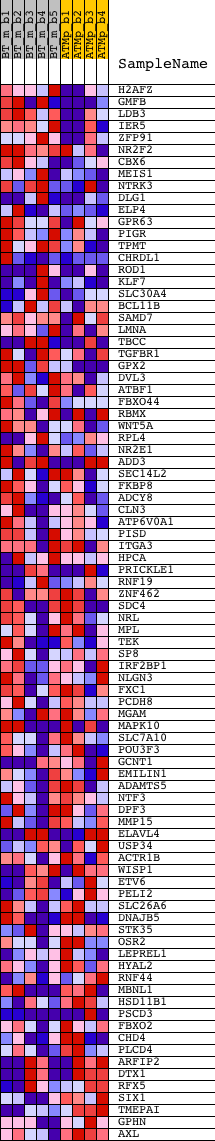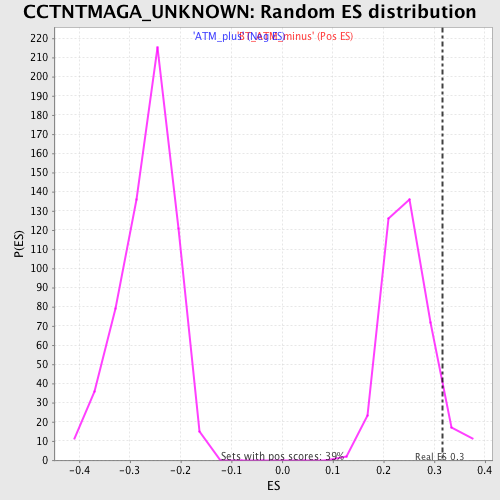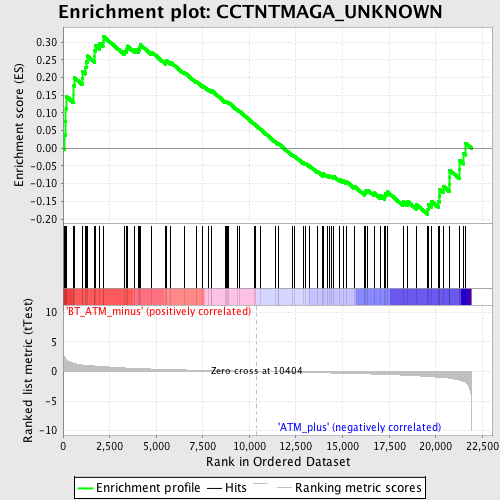
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | CCTNTMAGA_UNKNOWN |
| Enrichment Score (ES) | 0.31686148 |
| Normalized Enrichment Score (NES) | 1.2810054 |
| Nominal p-value | 0.062015504 |
| FDR q-value | 0.4802101 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | H2AFZ | 1441370_at | 92 | 2.365 | 0.0381 | Yes | ||
| 2 | GMFB | 1417069_a_at 1431686_a_at 1431687_at 1448570_at 1448571_a_at | 129 | 2.199 | 0.0758 | Yes | ||
| 3 | LDB3 | 1416752_at 1433783_at 1451999_at | 153 | 2.061 | 0.1116 | Yes | ||
| 4 | IER5 | 1417612_at 1417613_at | 171 | 1.992 | 0.1465 | Yes | ||
| 5 | ZFP91 | 1426326_at 1429615_at 1429616_at 1459444_at | 577 | 1.358 | 0.1523 | Yes | ||
| 6 | NR2F2 | 1416158_at 1416159_at 1416160_at 1431237_at 1444229_at | 578 | 1.356 | 0.1765 | Yes | ||
| 7 | CBX6 | 1424407_s_at 1429290_at | 609 | 1.337 | 0.1991 | Yes | ||
| 8 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 1047 | 1.075 | 0.1983 | Yes | ||
| 9 | NTRK3 | 1422329_a_at 1425070_at 1425071_s_at 1426003_at 1433825_at 1443970_at 1455917_at | 1060 | 1.071 | 0.2169 | Yes | ||
| 10 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 1217 | 1.027 | 0.2281 | Yes | ||
| 11 | ELP4 | 1416987_at 1447195_at 1448535_at | 1246 | 1.021 | 0.2451 | Yes | ||
| 12 | GPR63 | 1422338_at 1440725_at | 1296 | 1.010 | 0.2610 | Yes | ||
| 13 | PIGR | 1450060_at | 1684 | 0.928 | 0.2599 | Yes | ||
| 14 | TPMT | 1419121_at 1430889_a_at 1438087_at 1456273_x_at | 1686 | 0.928 | 0.2764 | Yes | ||
| 15 | CHRDL1 | 1421295_at 1424945_at 1434201_at 1456722_at | 1732 | 0.918 | 0.2908 | Yes | ||
| 16 | ROD1 | 1424083_at 1424084_at 1441407_at 1455819_at | 1964 | 0.865 | 0.2957 | Yes | ||
| 17 | KLF7 | 1419354_at 1419355_at 1419356_at 1449439_at 1458389_at | 2143 | 0.824 | 0.3023 | Yes | ||
| 18 | SLC30A4 | 1418843_at | 2147 | 0.823 | 0.3169 | Yes | ||
| 19 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 3305 | 0.595 | 0.2745 | No | ||
| 20 | SAMD7 | 1430817_at 1455782_at | 3402 | 0.578 | 0.2805 | No | ||
| 21 | LMNA | 1421654_a_at 1425472_a_at 1426868_x_at 1452212_at 1457670_s_at | 3441 | 0.572 | 0.2890 | No | ||
| 22 | TBCC | 1424644_at 1451423_at | 3857 | 0.515 | 0.2792 | No | ||
| 23 | TGFBR1 | 1420893_a_at 1420894_at 1420895_at 1446946_at | 4027 | 0.496 | 0.2803 | No | ||
| 24 | GPX2 | 1449279_at | 4119 | 0.482 | 0.2848 | No | ||
| 25 | DVL3 | 1420457_at | 4143 | 0.479 | 0.2923 | No | ||
| 26 | ATBF1 | 1420649_at 1420650_at 1429725_at 1445941_at 1446094_at 1449947_s_at 1453267_at 1460006_at | 4771 | 0.413 | 0.2710 | No | ||
| 27 | FBXO44 | 1430541_at 1434455_at | 5510 | 0.344 | 0.2434 | No | ||
| 28 | RBMX | 1416177_at 1416354_at 1416355_at 1426863_at 1437847_x_at | 5559 | 0.339 | 0.2473 | No | ||
| 29 | WNT5A | 1436791_at 1448818_at 1456976_at | 5790 | 0.319 | 0.2424 | No | ||
| 30 | RPL4 | 1425183_a_at | 6503 | 0.260 | 0.2145 | No | ||
| 31 | NR2E1 | 1434921_at 1457289_at | 7159 | 0.212 | 0.1883 | No | ||
| 32 | ADD3 | 1423297_at 1423298_at 1426574_a_at 1447077_at 1456162_x_at | 7495 | 0.189 | 0.1764 | No | ||
| 33 | SEC14L2 | 1424530_at | 7797 | 0.168 | 0.1656 | No | ||
| 34 | FKBP8 | 1416113_at | 7954 | 0.158 | 0.1613 | No | ||
| 35 | ADCY8 | 1418754_at 1443704_at | 7967 | 0.156 | 0.1635 | No | ||
| 36 | CLN3 | 1417551_at 1457789_at | 8705 | 0.107 | 0.1317 | No | ||
| 37 | ATP6V0A1 | 1417632_at 1425227_a_at 1459924_at 1460650_at | 8753 | 0.104 | 0.1314 | No | ||
| 38 | PISD | 1460585_x_at | 8830 | 0.100 | 0.1297 | No | ||
| 39 | ITGA3 | 1421997_s_at 1455158_at 1460305_at | 8891 | 0.096 | 0.1287 | No | ||
| 40 | HPCA | 1425833_a_at 1450930_at | 9348 | 0.067 | 0.1090 | No | ||
| 41 | PRICKLE1 | 1441663_at 1442172_at 1442400_at 1444759_at 1452249_at 1457637_at | 9454 | 0.061 | 0.1053 | No | ||
| 42 | RNF19 | 1417508_at 1417509_at | 10269 | 0.009 | 0.0682 | No | ||
| 43 | ZNF462 | 1439106_at 1456789_at | 10322 | 0.005 | 0.0660 | No | ||
| 44 | SDC4 | 1417654_at 1448793_a_at | 10620 | -0.013 | 0.0526 | No | ||
| 45 | NRL | 1450946_at | 11383 | -0.061 | 0.0188 | No | ||
| 46 | MPL | 1421461_at 1451919_a_at | 11548 | -0.071 | 0.0126 | No | ||
| 47 | TEK | 1418788_at 1444931_at | 11568 | -0.072 | 0.0130 | No | ||
| 48 | SP8 | 1440519_at | 12312 | -0.118 | -0.0189 | No | ||
| 49 | IRF2BP1 | 1451252_at | 12427 | -0.126 | -0.0219 | No | ||
| 50 | NLGN3 | 1456384_at | 12897 | -0.155 | -0.0406 | No | ||
| 51 | FXC1 | 1417916_a_at 1436416_x_at 1448887_x_at 1453461_at | 12996 | -0.161 | -0.0422 | No | ||
| 52 | PCDH8 | 1417051_at 1432491_at 1447825_x_at | 13234 | -0.176 | -0.0499 | No | ||
| 53 | MGAM | 1456777_at | 13667 | -0.206 | -0.0659 | No | ||
| 54 | MAPK10 | 1437195_x_at 1448342_at 1458002_at 1458062_at 1458513_at | 13942 | -0.224 | -0.0745 | No | ||
| 55 | SLC7A10 | 1421093_at | 13969 | -0.226 | -0.0716 | No | ||
| 56 | POU3F3 | 1422331_at 1435197_at | 14172 | -0.239 | -0.0766 | No | ||
| 57 | GCNT1 | 1460431_at | 14304 | -0.249 | -0.0781 | No | ||
| 58 | EMILIN1 | 1416414_at 1431834_a_at | 14428 | -0.257 | -0.0792 | No | ||
| 59 | ADAMTS5 | 1422561_at 1450658_at 1456404_at | 14541 | -0.266 | -0.0795 | No | ||
| 60 | NTF3 | 1434802_s_at 1450803_at | 14843 | -0.289 | -0.0881 | No | ||
| 61 | DPF3 | 1441659_at 1442686_at 1450162_at | 15034 | -0.302 | -0.0914 | No | ||
| 62 | MMP15 | 1422597_at 1437462_x_at 1439734_at | 15224 | -0.316 | -0.0944 | No | ||
| 63 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 15670 | -0.350 | -0.1085 | No | ||
| 64 | USP34 | 1434392_at 1434393_at 1444437_at 1446112_at 1460126_at | 16185 | -0.392 | -0.1250 | No | ||
| 65 | ACTR1B | 1426403_at 1447292_at 1447293_x_at 1452057_at | 16240 | -0.397 | -0.1204 | No | ||
| 66 | WISP1 | 1448593_at 1448594_at | 16359 | -0.407 | -0.1185 | No | ||
| 67 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 16696 | -0.437 | -0.1261 | No | ||
| 68 | PELI2 | 1419006_s_at 1436966_at 1437181_at | 17054 | -0.474 | -0.1339 | No | ||
| 69 | SLC26A6 | 1416275_at | 17282 | -0.497 | -0.1354 | No | ||
| 70 | DNAJB5 | 1421961_a_at 1421962_at 1450436_s_at | 17292 | -0.498 | -0.1269 | No | ||
| 71 | STK35 | 1431107_at 1439802_at | 17403 | -0.512 | -0.1228 | No | ||
| 72 | OSR2 | 1426155_a_at 1430950_at | 18255 | -0.617 | -0.1507 | No | ||
| 73 | LEPREL1 | 1436178_at 1444392_at | 18485 | -0.649 | -0.1496 | No | ||
| 74 | HYAL2 | 1448679_at | 18976 | -0.727 | -0.1590 | No | ||
| 75 | RNF44 | 1423532_at | 19588 | -0.839 | -0.1719 | No | ||
| 76 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 19632 | -0.847 | -0.1587 | No | ||
| 77 | HSD11B1 | 1449038_at | 19793 | -0.883 | -0.1503 | No | ||
| 78 | PSCD3 | 1418758_a_at 1431707_a_at 1446268_at | 20177 | -0.975 | -0.1504 | No | ||
| 79 | FBXO2 | 1427004_at | 20218 | -0.987 | -0.1345 | No | ||
| 80 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 20233 | -0.992 | -0.1174 | No | ||
| 81 | PLCD4 | 1437030_at | 20404 | -1.025 | -0.1069 | No | ||
| 82 | ARFIP2 | 1424240_at 1435498_at | 20747 | -1.131 | -0.1023 | No | ||
| 83 | DTX1 | 1425822_a_at 1458643_at | 20759 | -1.138 | -0.0824 | No | ||
| 84 | RFX5 | 1423103_at | 20763 | -1.139 | -0.0622 | No | ||
| 85 | SIX1 | 1427277_at | 21295 | -1.456 | -0.0604 | No | ||
| 86 | TMEPAI | 1422705_at | 21300 | -1.460 | -0.0345 | No | ||
| 87 | GPHN | 1426462_at 1426463_at 1430038_at 1444869_at 1445188_at 1458242_at | 21518 | -1.682 | -0.0143 | No | ||
| 88 | AXL | 1423586_at 1438621_x_at | 21622 | -1.861 | 0.0143 | No |