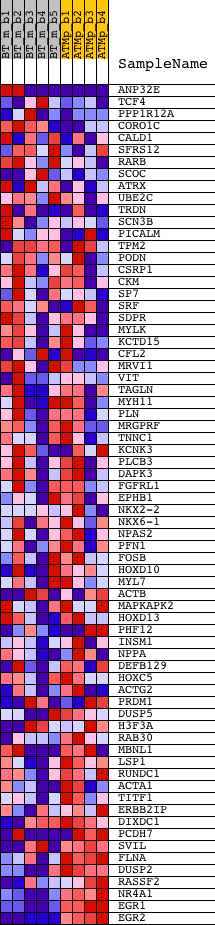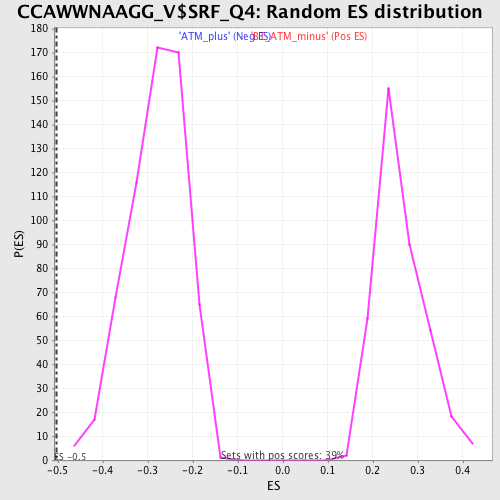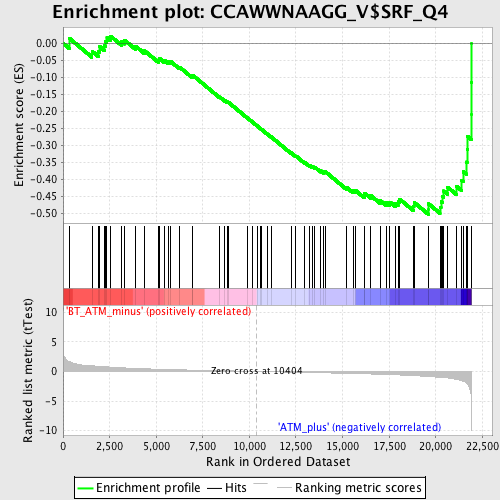
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CCAWWNAAGG_V$SRF_Q4 |
| Enrichment Score (ES) | -0.50284904 |
| Normalized Enrichment Score (NES) | -1.7950023 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025779774 |
| FWER p-Value | 0.093 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANP32E | 1420592_a_at 1447680_at | 345 | 1.629 | 0.0144 | No | ||
| 2 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 1551 | 0.958 | -0.0230 | No | ||
| 3 | PPP1R12A | 1429487_at 1437734_at 1437735_at 1444762_at 1444834_at 1453163_at | 1909 | 0.877 | -0.0231 | No | ||
| 4 | CORO1C | 1417752_at 1419911_at 1437721_at 1442972_at 1449660_s_at 1459169_at | 1968 | 0.864 | -0.0097 | No | ||
| 5 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 2233 | 0.801 | -0.0070 | No | ||
| 6 | SFRS12 | 1427134_at 1427135_at 1427136_s_at 1458056_at | 2287 | 0.790 | 0.0052 | No | ||
| 7 | RARB | 1454906_at | 2355 | 0.772 | 0.0165 | No | ||
| 8 | SCOC | 1416267_at 1430999_a_at | 2546 | 0.731 | 0.0213 | No | ||
| 9 | ATRX | 1420945_at 1420946_at 1420947_at 1438750_at 1441014_at 1441771_at 1450051_at 1453734_at 1456832_at | 3151 | 0.619 | 0.0052 | No | ||
| 10 | UBE2C | 1452954_at | 3308 | 0.593 | 0.0090 | No | ||
| 11 | TRDN | 1426134_at 1426142_a_at 1426143_at 1426144_x_at 1451801_at 1451940_x_at | 3892 | 0.510 | -0.0082 | No | ||
| 12 | SCN3B | 1426328_a_at 1435767_at | 4379 | 0.454 | -0.0220 | No | ||
| 13 | PICALM | 1441328_at 1443218_at 1446968_at 1451316_a_at 1455773_at | 5120 | 0.379 | -0.0489 | No | ||
| 14 | TPM2 | 1419738_a_at 1419739_at 1425028_a_at 1449577_x_at | 5160 | 0.375 | -0.0437 | No | ||
| 15 | PODN | 1435180_at | 5457 | 0.347 | -0.0508 | No | ||
| 16 | CSRP1 | 1425810_a_at 1425811_a_at | 5649 | 0.331 | -0.0534 | No | ||
| 17 | CKM | 1417614_at | 5769 | 0.321 | -0.0529 | No | ||
| 18 | SP7 | 1418425_at | 6247 | 0.280 | -0.0695 | No | ||
| 19 | SRF | 1418255_s_at 1418256_at | 6937 | 0.228 | -0.0968 | No | ||
| 20 | SDPR | 1416778_at 1416779_at 1443832_s_at | 6971 | 0.225 | -0.0942 | No | ||
| 21 | MYLK | 1425504_at 1425505_at 1425506_at | 8406 | 0.126 | -0.1574 | No | ||
| 22 | KCTD15 | 1435339_at | 8679 | 0.108 | -0.1678 | No | ||
| 23 | CFL2 | 1418066_at 1418067_at 1431432_at | 8810 | 0.101 | -0.1719 | No | ||
| 24 | MRVI1 | 1422245_a_at 1459665_s_at | 8868 | 0.098 | -0.1727 | No | ||
| 25 | VIT | 1426231_at | 9882 | 0.033 | -0.2184 | No | ||
| 26 | TAGLN | 1423505_at | 10169 | 0.015 | -0.2312 | No | ||
| 27 | MYH11 | 1418122_at 1448962_at | 10445 | -0.002 | -0.2438 | No | ||
| 28 | PLN | 1423359_at 1450952_at 1460332_at | 10580 | -0.010 | -0.2497 | No | ||
| 29 | MRGPRF | 1425894_at | 10677 | -0.016 | -0.2538 | No | ||
| 30 | TNNC1 | 1418370_at | 10996 | -0.038 | -0.2676 | No | ||
| 31 | KCNK3 | 1425341_at 1425342_a_at 1426058_a_at | 11167 | -0.048 | -0.2745 | No | ||
| 32 | PLCB3 | 1448661_at | 12247 | -0.113 | -0.3218 | No | ||
| 33 | DAPK3 | 1423446_at 1456496_at | 12501 | -0.130 | -0.3309 | No | ||
| 34 | FGFRL1 | 1447878_s_at 1451912_a_at | 12979 | -0.160 | -0.3498 | No | ||
| 35 | EPHB1 | 1455188_at | 13254 | -0.178 | -0.3590 | No | ||
| 36 | NKX2-2 | 1421112_at | 13380 | -0.186 | -0.3613 | No | ||
| 37 | NKX6-1 | 1425828_at | 13516 | -0.195 | -0.3639 | No | ||
| 38 | NPAS2 | 1421036_at 1421037_at | 13829 | -0.217 | -0.3741 | No | ||
| 39 | PFN1 | 1449018_at | 13989 | -0.227 | -0.3772 | No | ||
| 40 | FOSB | 1422134_at | 14066 | -0.232 | -0.3764 | No | ||
| 41 | HOXD10 | 1418606_at | 15240 | -0.318 | -0.4241 | No | ||
| 42 | MYL7 | 1449071_at | 15574 | -0.342 | -0.4330 | No | ||
| 43 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 15686 | -0.351 | -0.4316 | No | ||
| 44 | MAPKAPK2 | 1426648_at | 16184 | -0.392 | -0.4471 | No | ||
| 45 | HOXD13 | 1422239_at 1440626_at | 16192 | -0.393 | -0.4401 | No | ||
| 46 | PHF12 | 1434922_at 1453579_at | 16518 | -0.422 | -0.4472 | No | ||
| 47 | INSM1 | 1421399_at 1455865_at | 17027 | -0.471 | -0.4617 | No | ||
| 48 | NPPA | 1456062_at | 17376 | -0.509 | -0.4682 | No | ||
| 49 | DEFB129 | 1440957_at | 17548 | -0.527 | -0.4662 | No | ||
| 50 | HOXC5 | 1439885_at 1450832_at 1456301_at | 17858 | -0.564 | -0.4699 | No | ||
| 51 | ACTG2 | 1422340_a_at | 17989 | -0.581 | -0.4651 | No | ||
| 52 | PRDM1 | 1420425_at 1444997_at | 18069 | -0.593 | -0.4577 | No | ||
| 53 | DUSP5 | 1437199_at | 18806 | -0.699 | -0.4784 | No | ||
| 54 | H3F3A | 1423263_at 1442087_at | 18844 | -0.705 | -0.4671 | No | ||
| 55 | RAB30 | 1426452_a_at | 19627 | -0.847 | -0.4872 | Yes | ||
| 56 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 19632 | -0.847 | -0.4717 | Yes | ||
| 57 | LSP1 | 1417756_a_at | 20241 | -0.994 | -0.4811 | Yes | ||
| 58 | RUNDC1 | 1454829_at | 20295 | -1.003 | -0.4649 | Yes | ||
| 59 | ACTA1 | 1427735_a_at | 20375 | -1.018 | -0.4497 | Yes | ||
| 60 | TITF1 | 1422346_at | 20446 | -1.035 | -0.4337 | Yes | ||
| 61 | ERBB2IP | 1428011_a_at 1439079_a_at 1439080_at 1439638_at 1440573_at | 20654 | -1.094 | -0.4229 | Yes | ||
| 62 | DIXDC1 | 1425256_a_at 1435207_at 1443957_at 1444395_at | 21126 | -1.322 | -0.4200 | Yes | ||
| 63 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 21406 | -1.564 | -0.4038 | Yes | ||
| 64 | SVIL | 1427662_at 1460694_s_at 1460735_at | 21485 | -1.643 | -0.3769 | Yes | ||
| 65 | FLNA | 1426677_at | 21670 | -1.984 | -0.3486 | Yes | ||
| 66 | DUSP2 | 1450698_at | 21696 | -2.054 | -0.3117 | Yes | ||
| 67 | RASSF2 | 1428392_at 1444889_at | 21715 | -2.124 | -0.2732 | Yes | ||
| 68 | NR4A1 | 1416505_at | 21917 | -3.996 | -0.2084 | Yes | ||
| 69 | EGR1 | 1417065_at | 21922 | -4.993 | -0.1161 | Yes | ||
| 70 | EGR2 | 1427682_a_at 1427683_at | 21930 | -6.298 | 0.0002 | Yes |