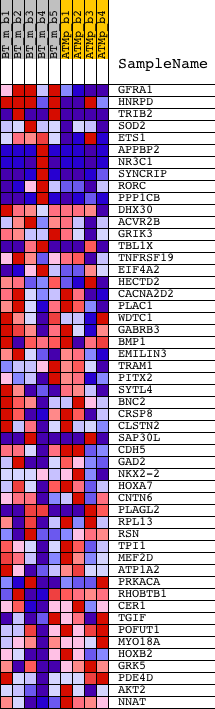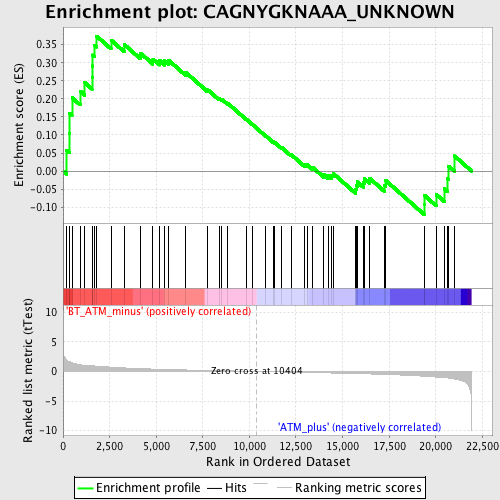
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | CAGNYGKNAAA_UNKNOWN |
| Enrichment Score (ES) | 0.3736998 |
| Normalized Enrichment Score (NES) | 1.3667803 |
| Nominal p-value | 0.051886793 |
| FDR q-value | 0.36152592 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GFRA1 | 1421973_at 1439015_at 1450440_at | 184 | 1.963 | 0.0575 | Yes | ||
| 2 | HNRPD | 1425142_a_at 1446083_at | 340 | 1.629 | 0.1051 | Yes | ||
| 3 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 346 | 1.628 | 0.1595 | Yes | ||
| 4 | SOD2 | 1417193_at 1417194_at 1442994_at 1444531_at 1448610_a_at 1454976_at | 479 | 1.475 | 0.2030 | Yes | ||
| 5 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 913 | 1.137 | 0.2214 | Yes | ||
| 6 | APPBP2 | 1430265_at 1434039_at 1434040_at 1434041_at 1444886_at 1447855_x_at 1451251_at | 1169 | 1.040 | 0.2447 | Yes | ||
| 7 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 1554 | 0.958 | 0.2593 | Yes | ||
| 8 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 1562 | 0.956 | 0.2910 | Yes | ||
| 9 | RORC | 1425792_a_at 1425793_a_at | 1595 | 0.950 | 0.3215 | Yes | ||
| 10 | PPP1CB | 1426205_at 1431328_at 1433540_x_at 1456462_x_at | 1678 | 0.930 | 0.3489 | Yes | ||
| 11 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 1799 | 0.902 | 0.3737 | Yes | ||
| 12 | ACVR2B | 1419140_at 1439856_at | 2574 | 0.726 | 0.3627 | No | ||
| 13 | GRIK3 | 1427709_at | 3275 | 0.598 | 0.3508 | No | ||
| 14 | TBL1X | 1434643_at 1434644_at 1441435_at 1446736_at 1455042_at | 4158 | 0.478 | 0.3265 | No | ||
| 15 | TNFRSF19 | 1415921_a_at 1425212_a_at 1427600_at 1445949_at 1448147_at | 4823 | 0.408 | 0.3099 | No | ||
| 16 | EIF4A2 | 1450934_at | 5178 | 0.373 | 0.3062 | No | ||
| 17 | HECTD2 | 1433944_at 1438596_at 1446224_at 1446430_at | 5460 | 0.347 | 0.3051 | No | ||
| 18 | CACNA2D2 | 1426185_at | 5664 | 0.330 | 0.3069 | No | ||
| 19 | PLAC1 | 1417553_at | 6586 | 0.254 | 0.2733 | No | ||
| 20 | WDTC1 | 1434560_at | 7775 | 0.170 | 0.2247 | No | ||
| 21 | GABRB3 | 1421189_at 1421190_at 1435021_at | 8380 | 0.128 | 0.2014 | No | ||
| 22 | BMP1 | 1426238_at 1427457_a_at 1427458_at 1446007_at | 8513 | 0.120 | 0.1994 | No | ||
| 23 | EMILIN3 | 1436965_at | 8826 | 0.100 | 0.1885 | No | ||
| 24 | TRAM1 | 1423732_at 1438225_x_at 1444506_at | 9836 | 0.037 | 0.1436 | No | ||
| 25 | PITX2 | 1424797_a_at 1450482_a_at | 10173 | 0.014 | 0.1287 | No | ||
| 26 | SYTL4 | 1417336_a_at 1425998_at | 10882 | -0.031 | 0.0974 | No | ||
| 27 | BNC2 | 1438861_at 1440593_at 1459132_at | 11271 | -0.055 | 0.0815 | No | ||
| 28 | CRSP8 | 1432462_a_at | 11368 | -0.060 | 0.0791 | No | ||
| 29 | CLSTN2 | 1422158_at 1441165_s_at 1442924_at | 11720 | -0.082 | 0.0659 | No | ||
| 30 | SAP30L | 1415718_at 1447379_at 1460071_at | 12245 | -0.113 | 0.0457 | No | ||
| 31 | CDH5 | 1422047_at 1433956_at | 12952 | -0.158 | 0.0188 | No | ||
| 32 | GAD2 | 1421978_at | 13103 | -0.167 | 0.0175 | No | ||
| 33 | NKX2-2 | 1421112_at | 13380 | -0.186 | 0.0112 | No | ||
| 34 | HOXA7 | 1449499_at | 13999 | -0.228 | -0.0094 | No | ||
| 35 | CNTN6 | 1422649_at | 14246 | -0.245 | -0.0125 | No | ||
| 36 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 14416 | -0.257 | -0.0116 | No | ||
| 37 | RPL13 | 1450150_a_at 1455245_x_at | 14505 | -0.263 | -0.0068 | No | ||
| 38 | RSN | 1425060_s_at 1431098_at 1439800_at 1444750_at 1450351_a_at 1452929_at | 15713 | -0.353 | -0.0501 | No | ||
| 39 | TPI1 | 1415918_a_at 1435659_a_at 1452927_x_at | 15741 | -0.354 | -0.0394 | No | ||
| 40 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 15790 | -0.359 | -0.0296 | No | ||
| 41 | ATP1A2 | 1427465_at 1434893_at 1443823_s_at 1452308_a_at 1455136_at | 16112 | -0.386 | -0.0313 | No | ||
| 42 | PRKACA | 1447720_x_at 1450519_a_at | 16164 | -0.390 | -0.0205 | No | ||
| 43 | RHOBTB1 | 1429206_at 1429656_at | 16465 | -0.418 | -0.0202 | No | ||
| 44 | CER1 | 1450256_at 1450257_at | 17247 | -0.494 | -0.0393 | No | ||
| 45 | TGIF | 1422286_a_at | 17305 | -0.499 | -0.0252 | No | ||
| 46 | POFUT1 | 1443881_at 1450137_at 1456323_at | 19386 | -0.798 | -0.0934 | No | ||
| 47 | MYO18A | 1451422_at | 19391 | -0.799 | -0.0668 | No | ||
| 48 | HOXB2 | 1444204_at 1449397_at | 20033 | -0.940 | -0.0645 | No | ||
| 49 | GRK5 | 1446361_at 1446998_at 1449514_at | 20463 | -1.039 | -0.0492 | No | ||
| 50 | PDE4D | 1443289_at 1458212_at 1459289_at 1459311_at | 20641 | -1.089 | -0.0208 | No | ||
| 51 | AKT2 | 1421324_a_at 1424480_s_at 1455703_at | 20705 | -1.113 | 0.0137 | No | ||
| 52 | NNAT | 1423506_a_at | 21018 | -1.263 | 0.0419 | No |