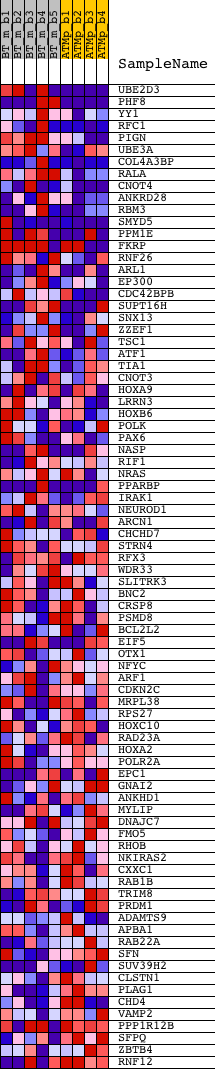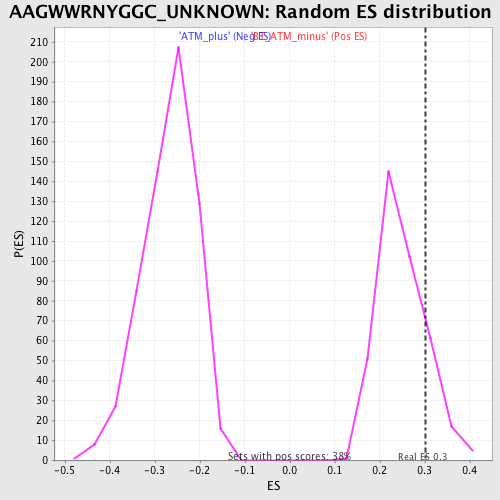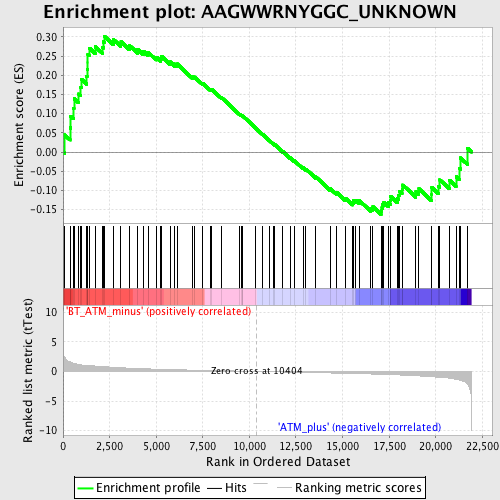
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | AAGWWRNYGGC_UNKNOWN |
| Enrichment Score (ES) | 0.30187988 |
| Normalized Enrichment Score (NES) | 1.2122582 |
| Nominal p-value | 0.16230367 |
| FDR q-value | 0.51511496 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 77 | 2.421 | 0.0449 | Yes | ||
| 2 | PHF8 | 1439132_at 1459392_at 1460398_at | 373 | 1.607 | 0.0635 | Yes | ||
| 3 | YY1 | 1422569_at 1422570_at 1435824_at 1457834_at | 413 | 1.567 | 0.0931 | Yes | ||
| 4 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 562 | 1.372 | 0.1137 | Yes | ||
| 5 | PIGN | 1421284_at 1425813_at 1425997_a_at 1432115_a_at | 587 | 1.352 | 0.1397 | Yes | ||
| 6 | UBE3A | 1416680_at 1416681_at 1416682_at 1425206_a_at 1431224_at 1445727_at | 846 | 1.170 | 0.1513 | Yes | ||
| 7 | COL4A3BP | 1420383_a_at 1420384_at 1449847_a_at 1452867_at | 950 | 1.121 | 0.1690 | Yes | ||
| 8 | RALA | 1423137_at 1450870_at 1456049_at | 988 | 1.097 | 0.1892 | Yes | ||
| 9 | CNOT4 | 1420999_at 1421000_at 1436645_a_at 1437586_at 1443169_at 1450083_at 1459449_at | 1263 | 1.017 | 0.1970 | Yes | ||
| 10 | ANKRD28 | 1434630_at 1454801_at | 1288 | 1.012 | 0.2161 | Yes | ||
| 11 | RBM3 | 1429169_at | 1330 | 1.003 | 0.2343 | Yes | ||
| 12 | SMYD5 | 1425479_at 1447749_at 1451722_s_at | 1331 | 1.003 | 0.2544 | Yes | ||
| 13 | PPM1E | 1434990_at 1445741_at | 1404 | 0.989 | 0.2709 | Yes | ||
| 14 | FKRP | 1436812_at 1437536_at 1438414_at | 1734 | 0.918 | 0.2742 | Yes | ||
| 15 | RNF26 | 1423650_at | 2112 | 0.831 | 0.2735 | Yes | ||
| 16 | ARL1 | 1447089_at 1447090_s_at 1451025_at | 2174 | 0.814 | 0.2870 | Yes | ||
| 17 | EP300 | 1434765_at | 2203 | 0.808 | 0.3019 | Yes | ||
| 18 | CDC42BPB | 1434652_at 1459167_at | 2698 | 0.701 | 0.2933 | No | ||
| 19 | SUPT16H | 1419741_at 1449578_at 1456449_at | 3107 | 0.625 | 0.2871 | No | ||
| 20 | SNX13 | 1435251_at 1444141_at 1454938_at | 3550 | 0.556 | 0.2780 | No | ||
| 21 | ZZEF1 | 1434192_at 1438691_at 1446081_at 1453583_at | 4006 | 0.499 | 0.2672 | No | ||
| 22 | TSC1 | 1422043_at 1439989_at 1455252_at | 4298 | 0.462 | 0.2631 | No | ||
| 23 | ATF1 | 1417296_at | 4559 | 0.434 | 0.2599 | No | ||
| 24 | TIA1 | 1416812_at 1416813_at 1416814_at 1423763_x_at 1431708_a_at 1437934_at 1441834_x_at 1453803_at 1454778_x_at | 5001 | 0.391 | 0.2475 | No | ||
| 25 | CNOT3 | 1435965_at 1441465_at | 5244 | 0.367 | 0.2438 | No | ||
| 26 | HOXA9 | 1421579_at 1455626_at | 5285 | 0.364 | 0.2492 | No | ||
| 27 | LRRN3 | 1434539_at | 5743 | 0.323 | 0.2348 | No | ||
| 28 | HOXB6 | 1451660_a_at | 5993 | 0.301 | 0.2294 | No | ||
| 29 | POLK | 1419501_at 1449483_at | 6118 | 0.291 | 0.2295 | No | ||
| 30 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 6943 | 0.227 | 0.1964 | No | ||
| 31 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 7057 | 0.218 | 0.1956 | No | ||
| 32 | RIF1 | 1438516_at 1445487_at | 7486 | 0.190 | 0.1798 | No | ||
| 33 | NRAS | 1422688_a_at 1454060_a_at | 7939 | 0.159 | 0.1623 | No | ||
| 34 | PPARBP | 1421906_at 1421907_at 1439408_a_at 1446250_at 1447450_at 1448708_at 1450402_at 1453972_x_at | 7991 | 0.155 | 0.1630 | No | ||
| 35 | IRAK1 | 1448668_a_at 1460649_at | 8492 | 0.121 | 0.1426 | No | ||
| 36 | NEUROD1 | 1426412_at 1426413_at | 9476 | 0.060 | 0.0988 | No | ||
| 37 | ARCN1 | 1423743_at 1436062_at 1444372_at 1451089_a_at | 9582 | 0.053 | 0.0950 | No | ||
| 38 | CHCHD7 | 1440158_x_at 1444318_at 1446844_at 1454640_at | 9626 | 0.050 | 0.0940 | No | ||
| 39 | STRN4 | 1426209_at | 9650 | 0.048 | 0.0940 | No | ||
| 40 | RFX3 | 1425413_at 1441253_at 1441702_at | 10357 | 0.003 | 0.0617 | No | ||
| 41 | WDR33 | 1418338_at 1423874_at 1444434_at 1444558_at 1453322_at 1453554_a_at 1455281_at 1456488_at 1456825_at 1458255_at | 10700 | -0.017 | 0.0464 | No | ||
| 42 | SLITRK3 | 1439250_at | 11068 | -0.042 | 0.0305 | No | ||
| 43 | BNC2 | 1438861_at 1440593_at 1459132_at | 11271 | -0.055 | 0.0223 | No | ||
| 44 | CRSP8 | 1432462_a_at | 11368 | -0.060 | 0.0191 | No | ||
| 45 | PSMD8 | 1423296_at | 11796 | -0.086 | 0.0013 | No | ||
| 46 | BCL2L2 | 1423572_at 1430453_a_at 1430454_x_at 1451029_at | 12203 | -0.111 | -0.0151 | No | ||
| 47 | EIF5 | 1415723_at 1433631_at 1454663_at 1454664_a_at 1456256_at | 12408 | -0.125 | -0.0219 | No | ||
| 48 | OTX1 | 1437601_at | 12888 | -0.155 | -0.0407 | No | ||
| 49 | NFYC | 1448963_at | 13041 | -0.163 | -0.0444 | No | ||
| 50 | ARF1 | 1420920_a_at 1426390_a_at | 13568 | -0.198 | -0.0645 | No | ||
| 51 | CDKN2C | 1416868_at 1439164_at | 14343 | -0.251 | -0.0949 | No | ||
| 52 | MRPL38 | 1430610_at 1444217_at 1447961_s_at | 14697 | -0.278 | -0.1055 | No | ||
| 53 | RPS27 | 1415716_a_at 1460699_at | 15142 | -0.310 | -0.1196 | No | ||
| 54 | HOXC10 | 1439798_at | 15542 | -0.340 | -0.1311 | No | ||
| 55 | RAD23A | 1422964_at 1437697_at 1453623_a_at 1455391_at | 15596 | -0.343 | -0.1266 | No | ||
| 56 | HOXA2 | 1419602_at | 15712 | -0.353 | -0.1249 | No | ||
| 57 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 15897 | -0.369 | -0.1259 | No | ||
| 58 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 16516 | -0.422 | -0.1457 | No | ||
| 59 | GNAI2 | 1419449_a_at 1435652_a_at | 16638 | -0.433 | -0.1426 | No | ||
| 60 | ANKHD1 | 1419867_a_at 1436597_at 1440754_at 1441597_at 1442641_at 1448008_at 1453023_at 1455759_a_at | 17074 | -0.477 | -0.1530 | No | ||
| 61 | MYLIP | 1424988_at | 17122 | -0.481 | -0.1455 | No | ||
| 62 | DNAJC7 | 1448362_at 1459704_at | 17142 | -0.483 | -0.1367 | No | ||
| 63 | FMO5 | 1421709_a_at 1440899_at 1450332_s_at | 17224 | -0.491 | -0.1306 | No | ||
| 64 | RHOB | 1449110_at | 17466 | -0.518 | -0.1313 | No | ||
| 65 | NKIRAS2 | 1424416_at | 17576 | -0.529 | -0.1257 | No | ||
| 66 | CXXC1 | 1452221_a_at 1454106_a_at | 17591 | -0.531 | -0.1157 | No | ||
| 67 | RAB1B | 1424064_at | 17953 | -0.577 | -0.1207 | No | ||
| 68 | TRIM8 | 1418577_at 1427593_at 1438679_at 1460237_at | 18027 | -0.587 | -0.1123 | No | ||
| 69 | PRDM1 | 1420425_at 1444997_at | 18069 | -0.593 | -0.1023 | No | ||
| 70 | ADAMTS9 | 1431399_at 1437785_at | 18201 | -0.609 | -0.0962 | No | ||
| 71 | APBA1 | 1437940_at 1455369_at 1459605_at | 18244 | -0.616 | -0.0858 | No | ||
| 72 | RAB22A | 1424503_at 1424504_at 1451379_at 1454988_s_at | 18946 | -0.720 | -0.1034 | No | ||
| 73 | SFN | 1448612_at | 19108 | -0.749 | -0.0958 | No | ||
| 74 | SUV39H2 | 1422979_at 1433996_at 1436561_at | 19794 | -0.883 | -0.1095 | No | ||
| 75 | CLSTN1 | 1421860_at 1421861_at 1445085_at | 19801 | -0.885 | -0.0921 | No | ||
| 76 | PLAG1 | 1421745_at 1443943_at | 20147 | -0.968 | -0.0885 | No | ||
| 77 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 20233 | -0.992 | -0.0726 | No | ||
| 78 | VAMP2 | 1420833_at 1420834_at | 20748 | -1.131 | -0.0735 | No | ||
| 79 | PPP1R12B | 1431462_at 1434786_at 1441730_at 1457169_at 1457338_at 1460091_at | 21103 | -1.310 | -0.0635 | No | ||
| 80 | SFPQ | 1423795_at 1423796_at 1436898_at 1438458_a_at 1438459_x_at 1439058_at | 21302 | -1.461 | -0.0433 | No | ||
| 81 | ZBTB4 | 1429722_at 1460488_at | 21344 | -1.502 | -0.0152 | No | ||
| 82 | RNF12 | 1417250_at 1418730_at 1418731_at 1439403_x_at 1440850_at 1443167_at | 21709 | -2.106 | 0.0103 | No |