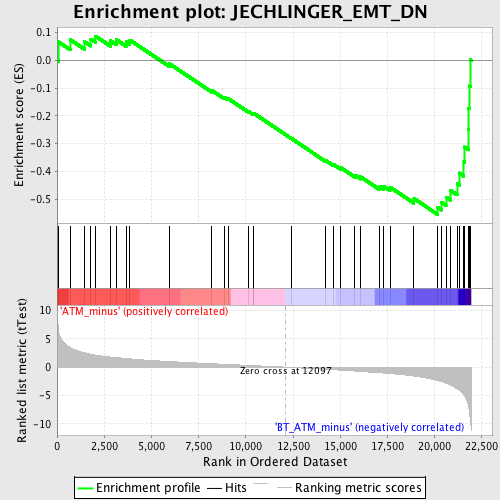
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | JECHLINGER_EMT_DN |
| Enrichment Score (ES) | -0.554408 |
| Normalized Enrichment Score (NES) | -1.7837628 |
| Nominal p-value | 0.0076045627 |
| FDR q-value | 0.116662905 |
| FWER p-Value | 0.894 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AMD1 | 1448484_at | 70 | 6.462 | 0.0658 | No | ||
| 2 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 702 | 3.417 | 0.0735 | No | ||
| 3 | FOSB | 1422134_at | 1438 | 2.551 | 0.0671 | No | ||
| 4 | ATF3 | 1449363_at | 1791 | 2.270 | 0.0753 | No | ||
| 5 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 2037 | 2.111 | 0.0866 | No | ||
| 6 | CTSH | 1418365_at 1443814_x_at | 2824 | 1.778 | 0.0697 | No | ||
| 7 | TACSTD1 | 1416579_a_at 1447899_x_at | 3129 | 1.682 | 0.0738 | No | ||
| 8 | ITGB5 | 1417533_a_at 1417534_at 1456133_x_at 1456195_x_at | 3653 | 1.520 | 0.0661 | No | ||
| 9 | TIAM1 | 1418057_at 1444373_at 1453887_a_at | 3855 | 1.464 | 0.0726 | No | ||
| 10 | SERPINB5 | 1421752_a_at 1424623_at 1438856_x_at 1441941_x_at | 5952 | 0.981 | -0.0127 | No | ||
| 11 | CTGF | 1416953_at | 8201 | 0.600 | -0.1089 | No | ||
| 12 | TIMP3 | 1419088_at 1419089_at 1449334_at 1449335_at | 8852 | 0.495 | -0.1333 | No | ||
| 13 | TGFB3 | 1417455_at | 9064 | 0.463 | -0.1380 | No | ||
| 14 | FBP2 | 1449088_at | 10141 | 0.301 | -0.1839 | No | ||
| 15 | VAMP8 | 1420624_a_at | 10395 | 0.261 | -0.1927 | No | ||
| 16 | F3 | 1417408_at | 10423 | 0.257 | -0.1912 | No | ||
| 17 | CDH1 | 1443509_at 1448261_at | 12386 | -0.050 | -0.2803 | No | ||
| 18 | HMMR | 1425815_a_at 1427541_x_at 1429871_at 1450156_a_at 1450157_a_at | 14220 | -0.365 | -0.3601 | No | ||
| 19 | KITLG | 1415854_at 1415855_at 1426152_a_at 1440621_at 1448117_at | 14612 | -0.441 | -0.3732 | No | ||
| 20 | NUMB | 1416891_at 1425368_a_at 1444337_at 1457175_at | 15008 | -0.525 | -0.3857 | No | ||
| 21 | GRB7 | 1448227_at | 15771 | -0.685 | -0.4131 | No | ||
| 22 | SGK | 1416041_at | 16074 | -0.755 | -0.4189 | No | ||
| 23 | ARHGEF1 | 1421164_a_at 1440403_at | 17096 | -0.998 | -0.4549 | No | ||
| 24 | TGM2 | 1417500_a_at 1426004_a_at 1433428_x_at 1437277_x_at 1438942_x_at 1455900_x_at | 17289 | -1.035 | -0.4526 | No | ||
| 25 | PKP1 | 1449586_at | 17632 | -1.123 | -0.4562 | No | ||
| 26 | NNT | 1416105_at 1432608_at 1456573_x_at | 18898 | -1.580 | -0.4971 | No | ||
| 27 | JUP | 1426873_s_at | 20154 | -2.387 | -0.5289 | Yes | ||
| 28 | ACTN4 | 1423449_a_at 1444258_at | 20362 | -2.553 | -0.5111 | Yes | ||
| 29 | IRF6 | 1418301_at | 20599 | -2.822 | -0.4918 | Yes | ||
| 30 | STAT5A | 1421469_a_at 1450259_a_at | 20852 | -3.200 | -0.4691 | Yes | ||
| 31 | FLNA | 1426677_at | 21187 | -3.814 | -0.4436 | Yes | ||
| 32 | ATP1A1 | 1423653_at 1435919_at 1435920_x_at 1451071_a_at 1458868_at | 21299 | -4.066 | -0.4053 | Yes | ||
| 33 | BCL6 | 1421818_at 1450381_a_at | 21546 | -4.892 | -0.3643 | Yes | ||
| 34 | RARA | 1450180_a_at | 21585 | -5.077 | -0.3118 | Yes | ||
| 35 | KLF2 | 1448890_at | 21774 | -6.770 | -0.2481 | Yes | ||
| 36 | EGR1 | 1417065_at | 21803 | -7.184 | -0.1727 | Yes | ||
| 37 | DUSP1 | 1448830_at | 21821 | -7.570 | -0.0926 | Yes | ||
| 38 | EGR2 | 1427682_a_at 1427683_at | 21881 | -9.155 | 0.0024 | Yes |

