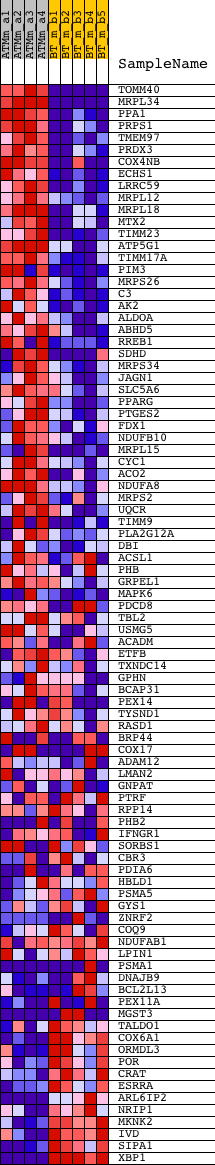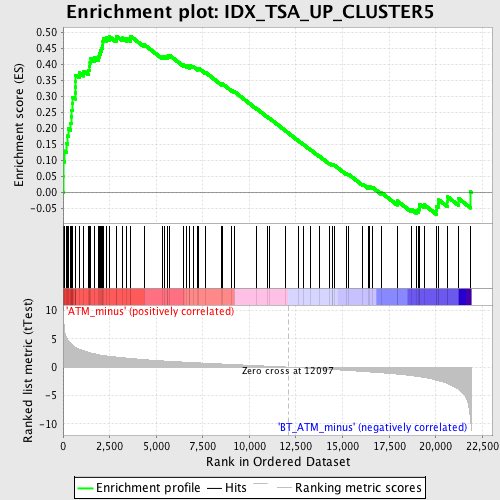
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | IDX_TSA_UP_CLUSTER5 |
| Enrichment Score (ES) | 0.48745927 |
| Normalized Enrichment Score (NES) | 1.9404372 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.01651254 |
| FWER p-Value | 0.203 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM40 | 1426118_a_at 1459680_at | 12 | 9.093 | 0.0487 | Yes | ||
| 2 | MRPL34 | 1416349_at 1438014_at | 17 | 9.026 | 0.0975 | Yes | ||
| 3 | PPA1 | 1416939_at | 74 | 6.371 | 0.1294 | Yes | ||
| 4 | PRPS1 | 1416052_at | 182 | 5.217 | 0.1528 | Yes | ||
| 5 | TMEM97 | 1416376_at 1440606_at | 239 | 4.835 | 0.1765 | Yes | ||
| 6 | PRDX3 | 1416292_at | 298 | 4.545 | 0.1984 | Yes | ||
| 7 | COX4NB | 1434952_at 1448677_at | 400 | 4.174 | 0.2164 | Yes | ||
| 8 | ECHS1 | 1452341_at | 434 | 4.051 | 0.2369 | Yes | ||
| 9 | LRRC59 | 1416234_at 1416235_at | 469 | 3.976 | 0.2569 | Yes | ||
| 10 | MRPL12 | 1452048_at | 495 | 3.920 | 0.2770 | Yes | ||
| 11 | MRPL18 | 1448373_at | 514 | 3.876 | 0.2972 | Yes | ||
| 12 | MTX2 | 1430500_s_at | 643 | 3.545 | 0.3105 | Yes | ||
| 13 | TIMM23 | 1416485_at | 657 | 3.507 | 0.3289 | Yes | ||
| 14 | ATP5G1 | 1416020_a_at 1444874_at | 666 | 3.489 | 0.3475 | Yes | ||
| 15 | TIMM17A | 1426256_at | 675 | 3.477 | 0.3660 | Yes | ||
| 16 | PIM3 | 1437100_x_at 1438956_x_at 1451069_at | 873 | 3.140 | 0.3740 | Yes | ||
| 17 | MRPS26 | 1433878_at | 1112 | 2.878 | 0.3787 | Yes | ||
| 18 | C3 | 1423954_at | 1368 | 2.614 | 0.3812 | Yes | ||
| 19 | AK2 | 1448450_at 1448451_at | 1416 | 2.576 | 0.3930 | Yes | ||
| 20 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 1441 | 2.546 | 0.4057 | Yes | ||
| 21 | ABHD5 | 1417565_at 1417566_at 1447548_at | 1491 | 2.500 | 0.4170 | Yes | ||
| 22 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 1672 | 2.355 | 0.4215 | Yes | ||
| 23 | SDHD | 1428235_at 1437489_x_at | 1894 | 2.196 | 0.4233 | Yes | ||
| 24 | MRPS34 | 1421971_a_at | 1954 | 2.156 | 0.4323 | Yes | ||
| 25 | JAGN1 | 1417064_at | 1995 | 2.132 | 0.4420 | Yes | ||
| 26 | SLC5A6 | 1435860_at | 2061 | 2.095 | 0.4504 | Yes | ||
| 27 | PPARG | 1420715_a_at | 2109 | 2.076 | 0.4595 | Yes | ||
| 28 | PTGES2 | 1417591_at | 2135 | 2.064 | 0.4695 | Yes | ||
| 29 | FDX1 | 1449108_at | 2145 | 2.050 | 0.4802 | Yes | ||
| 30 | NDUFB10 | 1428322_a_at | 2327 | 1.967 | 0.4826 | Yes | ||
| 31 | MRPL15 | 1425189_a_at 1430798_x_at 1451623_at 1460490_at | 2500 | 1.899 | 0.4850 | Yes | ||
| 32 | CYC1 | 1416604_at | 2845 | 1.769 | 0.4789 | Yes | ||
| 33 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 2867 | 1.764 | 0.4875 | Yes | ||
| 34 | NDUFA8 | 1423692_at | 3188 | 1.667 | 0.4818 | No | ||
| 35 | MRPS2 | 1420845_at 1420846_at | 3420 | 1.591 | 0.4799 | No | ||
| 36 | UQCR | 1448292_at | 3603 | 1.534 | 0.4799 | No | ||
| 37 | TIMM9 | 1428010_at 1449886_a_at | 3630 | 1.526 | 0.4870 | No | ||
| 38 | PLA2G12A | 1452026_a_at | 4343 | 1.330 | 0.4616 | No | ||
| 39 | DBI | 1422432_at 1433991_x_at 1438093_x_at 1455976_x_at | 5324 | 1.100 | 0.4227 | No | ||
| 40 | ACSL1 | 1422526_at 1423883_at 1447355_at 1450643_s_at 1460316_at | 5443 | 1.078 | 0.4231 | No | ||
| 41 | PHB | 1435144_at 1442233_at 1448563_at | 5589 | 1.049 | 0.4222 | No | ||
| 42 | GRPEL1 | 1417320_at | 5630 | 1.040 | 0.4260 | No | ||
| 43 | MAPK6 | 1419168_at 1419169_at 1429963_at 1459146_at | 5714 | 1.023 | 0.4277 | No | ||
| 44 | PDCD8 | 1418127_a_at | 6460 | 0.892 | 0.3984 | No | ||
| 45 | TBL2 | 1421927_at 1434224_at 1436603_at 1450412_at | 6635 | 0.861 | 0.3951 | No | ||
| 46 | USMG5 | 1448179_at | 6799 | 0.831 | 0.3922 | No | ||
| 47 | ACADM | 1415984_at | 6801 | 0.831 | 0.3967 | No | ||
| 48 | ETFB | 1428181_at | 6984 | 0.800 | 0.3927 | No | ||
| 49 | TXNDC14 | 1452130_at | 7226 | 0.758 | 0.3857 | No | ||
| 50 | GPHN | 1426462_at 1426463_at 1430038_at 1444869_at 1445188_at 1458242_at | 7272 | 0.752 | 0.3877 | No | ||
| 51 | BCAP31 | 1451049_at 1456279_a_at | 7662 | 0.688 | 0.3737 | No | ||
| 52 | PEX14 | 1419053_at 1458153_at | 8524 | 0.549 | 0.3372 | No | ||
| 53 | TYSND1 | 1428689_at 1428690_at 1441856_x_at | 8559 | 0.543 | 0.3386 | No | ||
| 54 | RASD1 | 1423619_at | 9051 | 0.465 | 0.3187 | No | ||
| 55 | BRP44 | 1423833_a_at 1430071_at | 9194 | 0.444 | 0.3146 | No | ||
| 56 | COX17 | 1434435_s_at 1434871_at | 10396 | 0.260 | 0.2610 | No | ||
| 57 | ADAM12 | 1421171_at 1421172_at | 10985 | 0.174 | 0.2350 | No | ||
| 58 | LMAN2 | 1423074_at 1423075_at | 11089 | 0.156 | 0.2312 | No | ||
| 59 | GNPAT | 1417456_at | 11954 | 0.021 | 0.1917 | No | ||
| 60 | PTRF | 1421431_at 1424130_a_at 1453891_at | 12665 | -0.101 | 0.1598 | No | ||
| 61 | RPP14 | 1419460_at 1419461_at 1443358_at | 12914 | -0.142 | 0.1492 | No | ||
| 62 | PHB2 | 1416202_at 1438938_x_at 1455713_x_at 1460035_at | 13268 | -0.202 | 0.1341 | No | ||
| 63 | IFNGR1 | 1443602_at 1448167_at | 13745 | -0.285 | 0.1139 | No | ||
| 64 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 14295 | -0.378 | 0.0908 | No | ||
| 65 | CBR3 | 1427912_at | 14446 | -0.407 | 0.0861 | No | ||
| 66 | PDIA6 | 1423648_at 1430983_at | 14470 | -0.414 | 0.0873 | No | ||
| 67 | HBLD1 | 1424154_a_at | 14564 | -0.431 | 0.0854 | No | ||
| 68 | PSMA5 | 1424681_a_at 1434356_a_at | 15216 | -0.567 | 0.0587 | No | ||
| 69 | GYS1 | 1450196_s_at | 15311 | -0.585 | 0.0575 | No | ||
| 70 | ZNRF2 | 1431154_at 1434016_at 1434017_at 1444735_at 1455290_at 1455291_s_at | 16082 | -0.757 | 0.0264 | No | ||
| 71 | COQ9 | 1428134_at 1440183_x_at 1445313_at | 16411 | -0.839 | 0.0159 | No | ||
| 72 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 16459 | -0.852 | 0.0184 | No | ||
| 73 | LPIN1 | 1418288_at 1426516_a_at | 16599 | -0.885 | 0.0168 | No | ||
| 74 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 17107 | -1.001 | -0.0010 | No | ||
| 75 | DNAJB9 | 1417191_at | 17940 | -1.215 | -0.0325 | No | ||
| 76 | BCL2L13 | 1424406_at 1429539_at | 17946 | -1.217 | -0.0261 | No | ||
| 77 | PEX11A | 1419365_at 1449442_at | 18691 | -1.493 | -0.0521 | No | ||
| 78 | MGST3 | 1448300_at 1450563_at | 18995 | -1.622 | -0.0571 | No | ||
| 79 | TALDO1 | 1425129_a_at | 19089 | -1.658 | -0.0524 | No | ||
| 80 | COX6A1 | 1417417_a_at | 19135 | -1.678 | -0.0454 | No | ||
| 81 | ORMDL3 | 1419450_at | 19147 | -1.684 | -0.0368 | No | ||
| 82 | POR | 1416933_at | 19394 | -1.798 | -0.0383 | No | ||
| 83 | CRAT | 1417008_at 1441919_x_at 1448544_at | 20054 | -2.299 | -0.0560 | No | ||
| 84 | ESRRA | 1442864_at 1460652_at | 20062 | -2.306 | -0.0438 | No | ||
| 85 | ARL6IP2 | 1416793_at 1416794_at 1431197_at 1435594_at | 20137 | -2.374 | -0.0343 | No | ||
| 86 | NRIP1 | 1418469_at 1432603_at 1434384_at 1446389_at 1447211_at 1449089_at 1454295_at | 20176 | -2.397 | -0.0231 | No | ||
| 87 | MKNK2 | 1418300_a_at 1449029_at | 20650 | -2.889 | -0.0291 | No | ||
| 88 | IVD | 1418238_at 1449001_at | 20656 | -2.896 | -0.0136 | No | ||
| 89 | SIPA1 | 1416206_at | 21252 | -3.966 | -0.0193 | No | ||
| 90 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 21887 | -9.319 | 0.0022 | No |