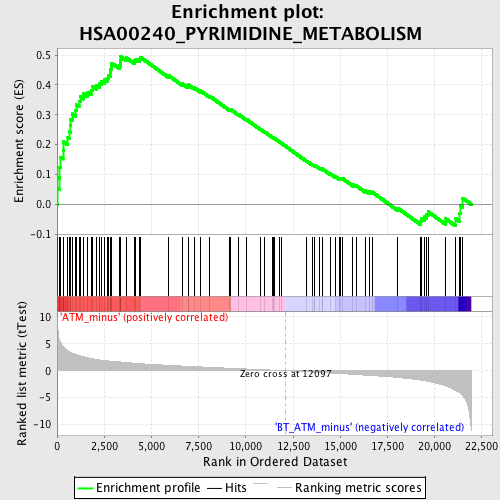
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | 0.49389622 |
| Normalized Enrichment Score (NES) | 1.9808749 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.019505257 |
| FWER p-Value | 0.112 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NT5C2 | 1417201_at 1425933_a_at 1442629_at 1443391_at 1448614_at | 38 | 8.230 | 0.0531 | Yes | ||
| 2 | POLR1A | 1417775_at 1446439_at 1447597_at 1447598_x_at 1456066_a_at 1460215_at | 107 | 5.837 | 0.0889 | Yes | ||
| 3 | CTPS | 1416563_at | 132 | 5.549 | 0.1248 | Yes | ||
| 4 | POLE3 | 1421015_s_at | 173 | 5.269 | 0.1581 | Yes | ||
| 5 | TXNRD2 | 1429971_at 1429972_s_at 1449097_at 1451936_a_at | 313 | 4.485 | 0.1817 | Yes | ||
| 6 | ITPA | 1416448_at 1442353_at | 343 | 4.353 | 0.2093 | Yes | ||
| 7 | NME4 | 1416798_a_at | 572 | 3.730 | 0.2238 | Yes | ||
| 8 | NME7 | 1418217_at 1438874_at | 660 | 3.505 | 0.2432 | Yes | ||
| 9 | POLR1D | 1447320_x_at 1448287_at 1451120_at 1459657_s_at | 692 | 3.445 | 0.2647 | Yes | ||
| 10 | POLR2K | 1452596_at | 728 | 3.373 | 0.2856 | Yes | ||
| 11 | DHODH | 1417581_at 1417582_s_at | 822 | 3.206 | 0.3027 | Yes | ||
| 12 | POLE | 1441854_at 1448650_a_at | 990 | 2.999 | 0.3151 | Yes | ||
| 13 | POLE4 | 1423371_at 1423372_at 1432286_at 1444739_at | 1032 | 2.964 | 0.3329 | Yes | ||
| 14 | DCTD | 1454659_at | 1163 | 2.819 | 0.3458 | Yes | ||
| 15 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 1231 | 2.751 | 0.3611 | Yes | ||
| 16 | NME2 | 1448808_a_at | 1415 | 2.576 | 0.3699 | Yes | ||
| 17 | CMPK | 1423073_at 1440308_at | 1627 | 2.389 | 0.3761 | Yes | ||
| 18 | AICDA | 1420577_at | 1840 | 2.241 | 0.3814 | Yes | ||
| 19 | NUDT2 | 1418737_at | 1871 | 2.218 | 0.3948 | Yes | ||
| 20 | UPB1 | 1460244_at | 2094 | 2.080 | 0.3985 | Yes | ||
| 21 | POLE2 | 1427094_at 1431440_at | 2258 | 1.996 | 0.4043 | Yes | ||
| 22 | ECGF1 | 1432181_s_at | 2356 | 1.956 | 0.4129 | Yes | ||
| 23 | TK1 | 1416258_at | 2530 | 1.884 | 0.4176 | Yes | ||
| 24 | POLR2L | 1428296_at | 2685 | 1.829 | 0.4227 | Yes | ||
| 25 | NT5M | 1453767_a_at | 2725 | 1.815 | 0.4331 | Yes | ||
| 26 | ENTPD4 | 1431761_at 1438177_x_at 1447900_x_at 1449190_a_at | 2822 | 1.779 | 0.4405 | Yes | ||
| 27 | NME6 | 1448574_at | 2829 | 1.775 | 0.4521 | Yes | ||
| 28 | UPP1 | 1448562_at | 2859 | 1.766 | 0.4625 | Yes | ||
| 29 | POLA2 | 1425794_at 1448369_at 1456713_at | 2897 | 1.755 | 0.4725 | Yes | ||
| 30 | POLR2B | 1433552_a_at | 3286 | 1.639 | 0.4657 | Yes | ||
| 31 | NP | 1416530_a_at | 3345 | 1.622 | 0.4739 | Yes | ||
| 32 | POLR2I | 1428494_a_at | 3373 | 1.611 | 0.4834 | Yes | ||
| 33 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 3378 | 1.608 | 0.4939 | Yes | ||
| 34 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 3670 | 1.514 | 0.4907 | No | ||
| 35 | POLR2D | 1424258_at | 4090 | 1.398 | 0.4808 | No | ||
| 36 | PNPT1 | 1444288_at 1452676_a_at | 4175 | 1.375 | 0.4861 | No | ||
| 37 | CAD | 1440857_at 1452829_at 1452830_s_at | 4380 | 1.320 | 0.4856 | No | ||
| 38 | UPP2 | 1460059_at | 4416 | 1.311 | 0.4927 | No | ||
| 39 | AK3 | 1423717_at 1423718_at 1432436_a_at 1458812_at | 5918 | 0.988 | 0.4306 | No | ||
| 40 | POLR3K | 1422752_at 1422753_a_at 1439266_a_at 1440735_at 1450737_at 1456731_x_at | 6627 | 0.862 | 0.4040 | No | ||
| 41 | DTYMK | 1438096_a_at 1449116_a_at 1452681_at | 6936 | 0.811 | 0.3953 | No | ||
| 42 | POLR2F | 1415754_at 1427402_at | 6937 | 0.810 | 0.4007 | No | ||
| 43 | POLR3G | 1449155_at | 7276 | 0.752 | 0.3902 | No | ||
| 44 | DPYD | 1427945_at 1427946_s_at 1443717_at | 7598 | 0.700 | 0.3802 | No | ||
| 45 | ENTPD8 | 1429550_at | 8081 | 0.618 | 0.3623 | No | ||
| 46 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 9110 | 0.456 | 0.3183 | No | ||
| 47 | POLR2G | 1416270_at | 9196 | 0.444 | 0.3173 | No | ||
| 48 | POLR3H | 1424227_at 1424228_at | 9626 | 0.382 | 0.3002 | No | ||
| 49 | CTPS2 | 1435759_at 1442998_at 1448111_at | 10031 | 0.316 | 0.2839 | No | ||
| 50 | POLR3B | 1443466_s_at 1443494_at 1452949_at 1457345_at | 10765 | 0.205 | 0.2517 | No | ||
| 51 | ENTPD6 | 1460433_at | 10992 | 0.173 | 0.2425 | No | ||
| 52 | CANT1 | 1421476_a_at 1433709_at 1451688_s_at | 11404 | 0.108 | 0.2244 | No | ||
| 53 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 11482 | 0.097 | 0.2215 | No | ||
| 54 | POLR2C | 1416341_at | 11509 | 0.093 | 0.2210 | No | ||
| 55 | NT5C3 | 1431306_at 1451050_at | 11794 | 0.046 | 0.2083 | No | ||
| 56 | POLR2H | 1424473_at | 11875 | 0.034 | 0.2048 | No | ||
| 57 | POLD1 | 1448187_at 1456055_x_at | 13226 | -0.194 | 0.1444 | No | ||
| 58 | NME1 | 1424110_a_at 1435277_x_at | 13512 | -0.242 | 0.1329 | No | ||
| 59 | UMPS | 1431652_at 1434859_at 1451445_at 1455832_a_at | 13626 | -0.264 | 0.1295 | No | ||
| 60 | DUT | 1419269_at 1419270_a_at | 13637 | -0.266 | 0.1308 | No | ||
| 61 | POLD2 | 1448277_at | 13894 | -0.306 | 0.1212 | No | ||
| 62 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 14031 | -0.331 | 0.1171 | No | ||
| 63 | DPYS | 1425688_a_at 1425689_at 1436291_a_at | 14044 | -0.334 | 0.1188 | No | ||
| 64 | CDA | 1427357_at | 14458 | -0.411 | 0.1027 | No | ||
| 65 | NT5C1B | 1427715_a_at | 14716 | -0.462 | 0.0940 | No | ||
| 66 | ENTPD5 | 1417382_at 1417383_at 1417384_at 1433763_at 1451765_a_at 1458677_at | 14952 | -0.511 | 0.0866 | No | ||
| 67 | RFC5 | 1452917_at | 15014 | -0.527 | 0.0873 | No | ||
| 68 | ENTPD1 | 1423326_at 1450939_at 1453586_at | 15094 | -0.543 | 0.0873 | No | ||
| 69 | NT5E | 1416517_at 1422974_at 1427379_at 1428547_at | 15666 | -0.665 | 0.0657 | No | ||
| 70 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 15830 | -0.697 | 0.0628 | No | ||
| 71 | POLR2J | 1417720_at 1440065_at | 16322 | -0.815 | 0.0458 | No | ||
| 72 | NT5C | 1417252_at | 16517 | -0.864 | 0.0427 | No | ||
| 73 | TXNRD1 | 1421529_a_at 1424486_a_at 1424487_x_at | 16680 | -0.909 | 0.0413 | No | ||
| 74 | ENTPD3 | 1434049_at | 18045 | -1.248 | -0.0128 | No | ||
| 75 | POLD4 | 1427885_at | 19246 | -1.730 | -0.0562 | No | ||
| 76 | RRM1 | 1415878_at 1440073_at 1448127_at | 19304 | -1.758 | -0.0470 | No | ||
| 77 | POLR3A | 1434453_at 1437525_a_at 1444310_at | 19469 | -1.841 | -0.0423 | No | ||
| 78 | TK2 | 1426100_a_at | 19570 | -1.898 | -0.0342 | No | ||
| 79 | RRM2B | 1433773_at 1437221_at 1437222_x_at 1437476_at | 19646 | -1.957 | -0.0246 | No | ||
| 80 | UCK1 | 1424399_at | 20584 | -2.808 | -0.0487 | No | ||
| 81 | DCK | 1428838_a_at 1439012_a_at 1449176_a_at | 21104 | -3.673 | -0.0480 | No | ||
| 82 | POLR3GL | 1451364_at | 21328 | -4.126 | -0.0307 | No | ||
| 83 | ZNRD1 | 1422517_a_at | 21343 | -4.168 | -0.0036 | No | ||
| 84 | UCK2 | 1417137_at 1426909_at 1439740_s_at 1439741_x_at 1441858_at 1448604_at 1457980_x_at | 21479 | -4.583 | 0.0208 | No |

