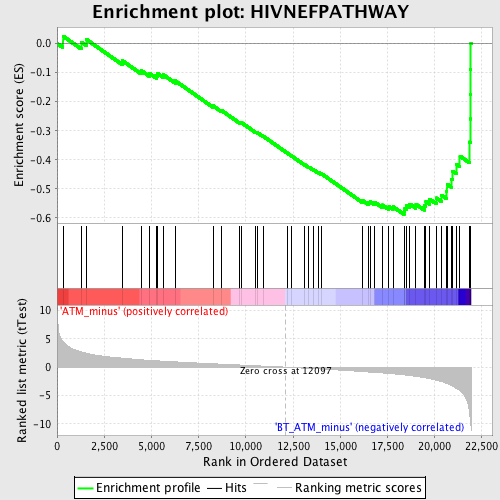
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | HIVNEFPATHWAY |
| Enrichment Score (ES) | -0.58793616 |
| Normalized Enrichment Score (NES) | -2.02853 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.021609621 |
| FWER p-Value | 0.063 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CYCS | 1422484_at 1445484_at | 311 | 4.490 | 0.0253 | No | ||
| 2 | CASP8 | 1424552_at | 1277 | 2.700 | 0.0049 | No | ||
| 3 | BID | 1417045_at 1447873_x_at 1448560_at | 1558 | 2.443 | 0.0136 | No | ||
| 4 | LMNB2 | 1451849_a_at | 3455 | 1.583 | -0.0591 | No | ||
| 5 | RASA1 | 1426476_at 1426477_at 1426478_at 1438998_at | 4454 | 1.298 | -0.0933 | No | ||
| 6 | PSEN2 | 1425869_a_at 1431542_at | 4889 | 1.188 | -0.1027 | No | ||
| 7 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 5266 | 1.113 | -0.1101 | No | ||
| 8 | TNF | 1419607_at | 5329 | 1.099 | -0.1033 | No | ||
| 9 | PSEN1 | 1420261_at 1421853_at 1425549_at 1450399_at | 5624 | 1.041 | -0.1076 | No | ||
| 10 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 6264 | 0.932 | -0.1286 | No | ||
| 11 | CDC2L1 | 1418841_s_at | 8255 | 0.590 | -0.2143 | No | ||
| 12 | LMNB1 | 1423520_at 1423521_at | 8697 | 0.521 | -0.2299 | No | ||
| 13 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 9681 | 0.373 | -0.2715 | No | ||
| 14 | PAK2 | 1434250_at 1454887_at | 9770 | 0.356 | -0.2724 | No | ||
| 15 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 10517 | 0.241 | -0.3044 | No | ||
| 16 | PRKCD | 1422847_a_at 1442256_at | 10621 | 0.228 | -0.3071 | No | ||
| 17 | NFKB1 | 1427705_a_at 1442949_at | 10904 | 0.186 | -0.3183 | No | ||
| 18 | RIPK1 | 1419508_at 1439273_at 1449485_at | 12208 | -0.017 | -0.3777 | No | ||
| 19 | LMNA | 1421654_a_at 1425472_a_at 1426868_x_at 1452212_at 1457670_s_at | 12432 | -0.058 | -0.3874 | No | ||
| 20 | CRADD | 1427505_a_at 1443430_at | 13107 | -0.175 | -0.4167 | No | ||
| 21 | TNFRSF1B | 1418099_at 1448951_at | 13336 | -0.213 | -0.4252 | No | ||
| 22 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 13566 | -0.252 | -0.4335 | No | ||
| 23 | NUMA1 | 1423974_at 1423975_s_at 1441090_at 1444576_at 1444577_x_at | 13832 | -0.297 | -0.4430 | No | ||
| 24 | GSN | 1415812_at 1436991_x_at 1437171_x_at 1441225_at 1456312_x_at 1456568_at 1456569_x_at | 14015 | -0.327 | -0.4484 | No | ||
| 25 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 16164 | -0.778 | -0.5397 | No | ||
| 26 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 16482 | -0.856 | -0.5467 | No | ||
| 27 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 16585 | -0.880 | -0.5436 | No | ||
| 28 | TRADD | 1429117_at 1439910_a_at 1452622_a_at | 16821 | -0.940 | -0.5461 | No | ||
| 29 | BAG4 | 1418707_at 1439024_at 1449186_at | 17219 | -1.023 | -0.5552 | No | ||
| 30 | BIRC3 | 1421392_a_at 1425223_at | 17574 | -1.109 | -0.5616 | No | ||
| 31 | DAXX | 1419026_at | 17792 | -1.168 | -0.5613 | No | ||
| 32 | APAF1 | 1450223_at | 18376 | -1.358 | -0.5760 | Yes | ||
| 33 | CASP9 | 1426125_a_at 1437537_at 1457816_at | 18419 | -1.374 | -0.5658 | Yes | ||
| 34 | FADD | 1416888_at 1446499_at | 18521 | -1.412 | -0.5580 | Yes | ||
| 35 | CFLAR | 1424996_at 1425686_at 1425687_at 1439600_at 1449317_at | 18673 | -1.483 | -0.5519 | Yes | ||
| 36 | CASP2 | 1448165_at | 19003 | -1.624 | -0.5526 | Yes | ||
| 37 | BIRC2 | 1418854_at | 19444 | -1.821 | -0.5567 | Yes | ||
| 38 | MAP3K14 | 1422999_at 1434364_at | 19532 | -1.872 | -0.5443 | Yes | ||
| 39 | TRAF1 | 1423602_at 1445452_at | 19721 | -2.020 | -0.5351 | Yes | ||
| 40 | PRKDC | 1451576_at | 20082 | -2.318 | -0.5312 | Yes | ||
| 41 | BIRC4 | 1421394_a_at 1426636_a_at 1437533_at 1440351_at 1450231_a_at 1450232_at 1456088_at | 20353 | -2.545 | -0.5211 | Yes | ||
| 42 | ACTG1 | 1415779_s_at | 20639 | -2.878 | -0.5088 | Yes | ||
| 43 | MAP3K1 | 1424850_at | 20648 | -2.888 | -0.4838 | Yes | ||
| 44 | RELA | 1419536_a_at | 20889 | -3.265 | -0.4661 | Yes | ||
| 45 | DFFB | 1421229_at 1437051_at | 20947 | -3.380 | -0.4390 | Yes | ||
| 46 | ARHGDIB | 1426454_at | 21139 | -3.736 | -0.4148 | Yes | ||
| 47 | TRAF2 | 1451233_at | 21335 | -4.141 | -0.3873 | Yes | ||
| 48 | TNFRSF1A | 1417291_at | 21839 | -8.146 | -0.3387 | Yes | ||
| 49 | CASP7 | 1426062_a_at 1448659_at | 21883 | -9.234 | -0.2595 | Yes | ||
| 50 | CASP6 | 1415995_at 1458715_at | 21892 | -9.592 | -0.1755 | Yes | ||
| 51 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 21902 | -9.953 | -0.0884 | Yes | ||
| 52 | DFFA | 1450885_at 1457564_at | 21915 | -10.213 | 0.0009 | Yes |

