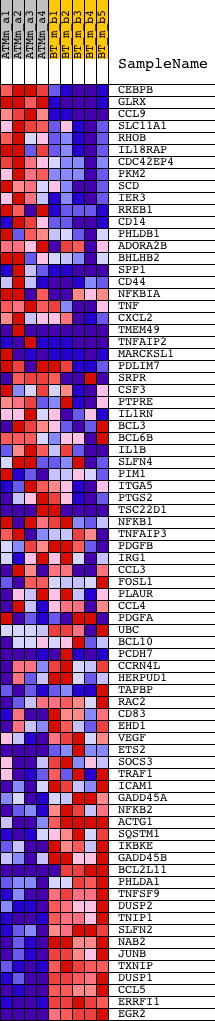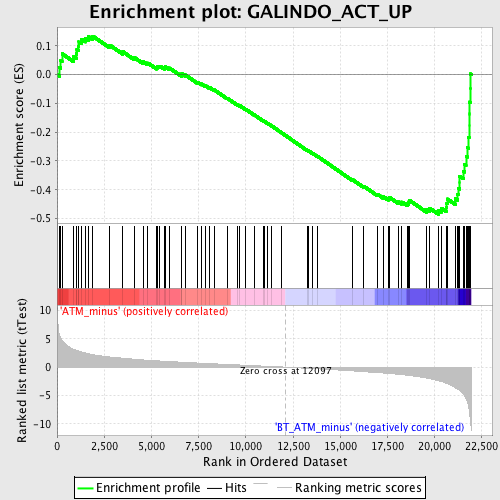
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GALINDO_ACT_UP |
| Enrichment Score (ES) | -0.48582652 |
| Normalized Enrichment Score (NES) | -1.8094395 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.112734444 |
| FWER p-Value | 0.809 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 1418901_at 1427844_a_at | 129 | 5.556 | 0.0247 | No | ||
| 2 | GLRX | 1416592_at 1416593_at | 196 | 5.110 | 0.0498 | No | ||
| 3 | CCL9 | 1417936_at 1448898_at | 263 | 4.732 | 0.0728 | No | ||
| 4 | SLC11A1 | 1420361_at | 866 | 3.150 | 0.0626 | No | ||
| 5 | RHOB | 1449110_at | 1015 | 2.977 | 0.0723 | No | ||
| 6 | IL18RAP | 1421291_at 1456545_at | 1048 | 2.947 | 0.0870 | No | ||
| 7 | CDC42EP4 | 1416511_a_at 1436801_x_at | 1125 | 2.863 | 0.0993 | No | ||
| 8 | PKM2 | 1417308_at | 1155 | 2.826 | 0.1135 | No | ||
| 9 | SCD | 1415964_at 1415965_at | 1293 | 2.680 | 0.1220 | No | ||
| 10 | IER3 | 1419647_a_at | 1484 | 2.505 | 0.1271 | No | ||
| 11 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 1672 | 2.355 | 0.1315 | No | ||
| 12 | CD14 | 1417268_at | 1889 | 2.203 | 0.1338 | No | ||
| 13 | PHLDB1 | 1424467_at 1424468_s_at | 2788 | 1.792 | 0.1026 | No | ||
| 14 | ADORA2B | 1434430_s_at 1434431_x_at 1434772_at 1450214_at | 3482 | 1.571 | 0.0795 | No | ||
| 15 | BHLHB2 | 1418025_at | 4094 | 1.397 | 0.0592 | No | ||
| 16 | SPP1 | 1449254_at | 4556 | 1.268 | 0.0451 | No | ||
| 17 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 4805 | 1.206 | 0.0404 | No | ||
| 18 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 5266 | 1.113 | 0.0255 | No | ||
| 19 | TNF | 1419607_at | 5329 | 1.099 | 0.0287 | No | ||
| 20 | CXCL2 | 1449984_at | 5436 | 1.080 | 0.0298 | No | ||
| 21 | TMEM49 | 1421491_a_at 1423722_at 1444820_at | 5669 | 1.033 | 0.0249 | No | ||
| 22 | TNFAIP2 | 1416273_at 1438855_x_at 1457918_at | 5726 | 1.021 | 0.0280 | No | ||
| 23 | MARCKSL1 | 1415922_s_at 1435415_x_at 1437226_x_at | 5951 | 0.982 | 0.0231 | No | ||
| 24 | PDLIM7 | 1417959_at 1428319_at 1443785_x_at | 6604 | 0.868 | -0.0019 | No | ||
| 25 | SRPR | 1423670_a_at 1427570_at | 6607 | 0.867 | 0.0027 | No | ||
| 26 | CSF3 | 1419427_at | 6792 | 0.833 | -0.0011 | No | ||
| 27 | PTPRE | 1418539_a_at 1418540_a_at 1459638_at | 7421 | 0.728 | -0.0258 | No | ||
| 28 | IL1RN | 1423017_a_at 1425663_at 1451798_at | 7646 | 0.691 | -0.0323 | No | ||
| 29 | BCL3 | 1418133_at | 7849 | 0.658 | -0.0379 | No | ||
| 30 | BCL6B | 1418421_at | 8072 | 0.619 | -0.0446 | No | ||
| 31 | IL1B | 1449399_a_at | 8326 | 0.582 | -0.0530 | No | ||
| 32 | SLFN4 | 1427102_at | 9015 | 0.472 | -0.0819 | No | ||
| 33 | PIM1 | 1423006_at 1435458_at | 9576 | 0.389 | -0.1054 | No | ||
| 34 | ITGA5 | 1423267_s_at 1423268_at 1457561_at 1458996_at | 9682 | 0.373 | -0.1081 | No | ||
| 35 | PTGS2 | 1417262_at 1417263_at | 9972 | 0.326 | -0.1195 | No | ||
| 36 | TSC22D1 | 1425742_a_at 1433899_x_at 1447360_at 1454758_a_at 1454971_x_at 1456132_x_at | 10444 | 0.254 | -0.1397 | No | ||
| 37 | NFKB1 | 1427705_a_at 1442949_at | 10904 | 0.186 | -0.1597 | No | ||
| 38 | TNFAIP3 | 1433699_at 1450829_at | 10976 | 0.174 | -0.1620 | No | ||
| 39 | PDGFB | 1450413_at 1450414_at | 11145 | 0.147 | -0.1688 | No | ||
| 40 | IRG1 | 1427381_at | 11353 | 0.115 | -0.1777 | No | ||
| 41 | CCL3 | 1419561_at | 11887 | 0.032 | -0.2019 | No | ||
| 42 | FOSL1 | 1417487_at 1417488_at | 13244 | -0.198 | -0.2628 | No | ||
| 43 | PLAUR | 1452521_a_at | 13317 | -0.210 | -0.2650 | No | ||
| 44 | CCL4 | 1421578_at | 13524 | -0.243 | -0.2730 | No | ||
| 45 | PDGFA | 1418711_at 1449187_at | 13771 | -0.289 | -0.2827 | No | ||
| 46 | UBC | 1420494_x_at 1425965_at 1425966_x_at 1432827_x_at 1437666_x_at 1454373_x_at | 15623 | -0.656 | -0.3638 | No | ||
| 47 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 16226 | -0.792 | -0.3870 | No | ||
| 48 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 16972 | -0.972 | -0.4157 | No | ||
| 49 | CCRN4L | 1425837_a_at 1458699_at | 17268 | -1.032 | -0.4235 | No | ||
| 50 | HERPUD1 | 1435626_a_at 1448185_at | 17537 | -1.101 | -0.4297 | No | ||
| 51 | TAPBP | 1421812_at 1450378_at | 17595 | -1.113 | -0.4262 | No | ||
| 52 | RAC2 | 1417620_at 1440208_at | 18088 | -1.263 | -0.4417 | No | ||
| 53 | CD83 | 1416111_at | 18263 | -1.319 | -0.4425 | No | ||
| 54 | EHD1 | 1416010_a_at 1416011_x_at 1416012_at 1448175_at | 18553 | -1.426 | -0.4478 | No | ||
| 55 | VEGF | 1420909_at 1451959_a_at | 18611 | -1.453 | -0.4424 | No | ||
| 56 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 18658 | -1.475 | -0.4364 | No | ||
| 57 | SOCS3 | 1416576_at 1455899_x_at 1456212_x_at | 19575 | -1.903 | -0.4678 | No | ||
| 58 | TRAF1 | 1423602_at 1445452_at | 19721 | -2.020 | -0.4634 | No | ||
| 59 | ICAM1 | 1424067_at | 20213 | -2.427 | -0.4725 | Yes | ||
| 60 | GADD45A | 1449519_at | 20336 | -2.523 | -0.4642 | Yes | ||
| 61 | NFKB2 | 1425902_a_at 1429128_x_at | 20604 | -2.837 | -0.4607 | Yes | ||
| 62 | ACTG1 | 1415779_s_at | 20639 | -2.878 | -0.4465 | Yes | ||
| 63 | SQSTM1 | 1440076_at 1442237_at 1444021_at 1450957_a_at | 20674 | -2.919 | -0.4319 | Yes | ||
| 64 | IKBKE | 1417813_at | 21092 | -3.638 | -0.4310 | Yes | ||
| 65 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21203 | -3.850 | -0.4148 | Yes | ||
| 66 | BCL2L11 | 1426334_a_at 1435448_at 1435449_at 1456005_a_at 1456006_at 1459794_at | 21270 | -4.007 | -0.3958 | Yes | ||
| 67 | PHLDA1 | 1418835_at | 21312 | -4.094 | -0.3751 | Yes | ||
| 68 | TNFSF9 | 1422924_at | 21314 | -4.099 | -0.3526 | Yes | ||
| 69 | DUSP2 | 1450698_at | 21513 | -4.754 | -0.3355 | Yes | ||
| 70 | TNIP1 | 1427689_a_at | 21579 | -5.049 | -0.3107 | Yes | ||
| 71 | SLFN2 | 1450165_at | 21657 | -5.545 | -0.2837 | Yes | ||
| 72 | NAB2 | 1417930_at | 21717 | -6.076 | -0.2529 | Yes | ||
| 73 | JUNB | 1415899_at | 21777 | -6.806 | -0.2181 | Yes | ||
| 74 | TXNIP | 1415996_at 1415997_at | 21820 | -7.564 | -0.1784 | Yes | ||
| 75 | DUSP1 | 1448830_at | 21821 | -7.570 | -0.1367 | Yes | ||
| 76 | CCL5 | 1418126_at | 21823 | -7.580 | -0.0950 | Yes | ||
| 77 | ERRFI1 | 1416129_at 1419816_s_at | 21877 | -9.011 | -0.0478 | Yes | ||
| 78 | EGR2 | 1427682_a_at 1427683_at | 21881 | -9.155 | 0.0024 | Yes |