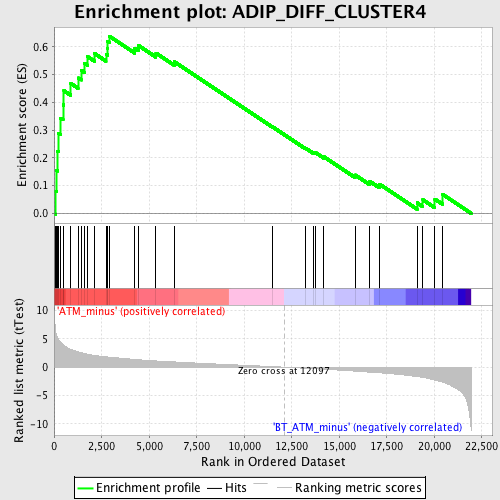
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | ADIP_DIFF_CLUSTER4 |
| Enrichment Score (ES) | 0.638747 |
| Normalized Enrichment Score (NES) | 2.078017 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009805887 |
| FWER p-Value | 0.02 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDX21 | 1448270_at 1448271_a_at | 86 | 6.176 | 0.0804 | Yes | ||
| 2 | MCM4 | 1416214_at 1436708_x_at | 124 | 5.592 | 0.1551 | Yes | ||
| 3 | PRPS1 | 1416052_at | 182 | 5.217 | 0.2237 | Yes | ||
| 4 | ECM1 | 1448613_at | 222 | 4.912 | 0.2890 | Yes | ||
| 5 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 351 | 4.320 | 0.3422 | Yes | ||
| 6 | IFRD2 | 1451016_at | 471 | 3.976 | 0.3910 | Yes | ||
| 7 | MRPL18 | 1448373_at | 514 | 3.876 | 0.4420 | Yes | ||
| 8 | RAD51 | 1418281_at | 879 | 3.127 | 0.4681 | Yes | ||
| 9 | ABCF2 | 1423863_at | 1260 | 2.724 | 0.4879 | Yes | ||
| 10 | ASS1 | 1416239_at 1459937_at | 1422 | 2.571 | 0.5157 | Yes | ||
| 11 | HAT1 | 1428061_at | 1588 | 2.416 | 0.5412 | Yes | ||
| 12 | CDCA7 | 1428069_at 1445681_at | 1753 | 2.296 | 0.5650 | Yes | ||
| 13 | NAP1L4 | 1448476_at | 2123 | 2.070 | 0.5764 | Yes | ||
| 14 | SAA2 | 1449326_x_at | 2730 | 1.814 | 0.5735 | Yes | ||
| 15 | CHAF1B | 1423877_at 1431275_at | 2803 | 1.787 | 0.5946 | Yes | ||
| 16 | HMGA1 | 1416184_s_at | 2813 | 1.783 | 0.6186 | Yes | ||
| 17 | POLA2 | 1425794_at 1448369_at 1456713_at | 2897 | 1.755 | 0.6387 | Yes | ||
| 18 | SNX5 | 1417646_a_at 1417647_at 1417648_s_at 1437074_at 1448791_at | 4241 | 1.354 | 0.5959 | No | ||
| 19 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 4450 | 1.300 | 0.6042 | No | ||
| 20 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 5354 | 1.095 | 0.5779 | No | ||
| 21 | ABCF1 | 1420157_s_at 1420158_s_at 1426921_at 1427444_at 1452236_at 1452429_s_at | 6315 | 0.920 | 0.5466 | No | ||
| 22 | SMC2 | 1429658_a_at 1429659_at 1429660_s_at 1448635_at 1458479_at | 11490 | 0.096 | 0.3117 | No | ||
| 23 | POLD1 | 1448187_at 1456055_x_at | 13226 | -0.194 | 0.2351 | No | ||
| 24 | DUT | 1419269_at 1419270_a_at | 13637 | -0.266 | 0.2200 | No | ||
| 25 | DNMT1 | 1422946_a_at 1435122_x_at 1445637_at 1447877_x_at | 13764 | -0.288 | 0.2182 | No | ||
| 26 | HELLS | 1417541_at 1430139_at 1453361_at | 14192 | -0.359 | 0.2036 | No | ||
| 27 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 15830 | -0.697 | 0.1384 | No | ||
| 28 | FEN1 | 1421731_a_at 1436454_x_at | 16603 | -0.886 | 0.1153 | No | ||
| 29 | INTS2 | 1429461_at 1436247_at | 17116 | -1.003 | 0.1056 | No | ||
| 30 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 19093 | -1.661 | 0.0380 | No | ||
| 31 | MCM5 | 1415945_at 1436808_x_at | 19397 | -1.799 | 0.0488 | No | ||
| 32 | ASF1B | 1423714_at | 20028 | -2.280 | 0.0511 | No | ||
| 33 | CDC6 | 1417019_a_at | 20444 | -2.626 | 0.0680 | No |

