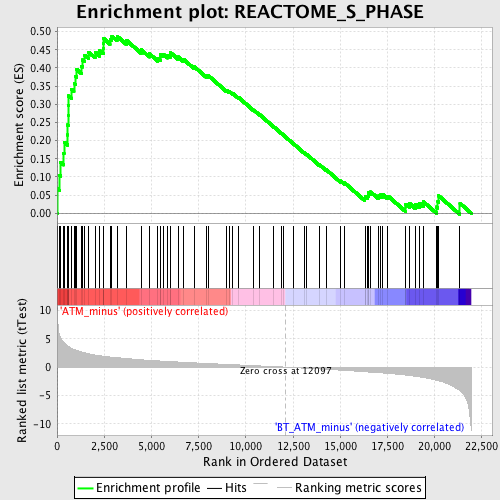
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_S_PHASE |
| Enrichment Score (ES) | 0.48762208 |
| Normalized Enrichment Score (NES) | 1.8893301 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.036325008 |
| FWER p-Value | 0.227 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC25A | 1417131_at 1417132_at 1445995_at | 8 | 9.163 | 0.0680 | Yes | ||
| 2 | MCM4 | 1416214_at 1436708_x_at | 124 | 5.592 | 0.1044 | Yes | ||
| 3 | PSMD12 | 1448492_a_at | 186 | 5.191 | 0.1403 | Yes | ||
| 4 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 351 | 4.320 | 0.1650 | Yes | ||
| 5 | PSMA2 | 1448206_at | 369 | 4.265 | 0.1960 | Yes | ||
| 6 | MNAT1 | 1432177_a_at | 556 | 3.770 | 0.2156 | Yes | ||
| 7 | PSMA3 | 1448442_a_at | 565 | 3.743 | 0.2432 | Yes | ||
| 8 | PSMC6 | 1417771_a_at | 588 | 3.680 | 0.2696 | Yes | ||
| 9 | PSMB10 | 1448632_at | 596 | 3.662 | 0.2966 | Yes | ||
| 10 | PSMD5 | 1423234_at | 608 | 3.626 | 0.3231 | Yes | ||
| 11 | RFC3 | 1423700_at 1432538_a_at | 752 | 3.338 | 0.3415 | Yes | ||
| 12 | PSMA4 | 1460339_at | 895 | 3.107 | 0.3581 | Yes | ||
| 13 | POLE | 1441854_at 1448650_a_at | 990 | 2.999 | 0.3762 | Yes | ||
| 14 | PSMC1 | 1416005_at | 1040 | 2.957 | 0.3960 | Yes | ||
| 15 | CDK4 | 1422441_x_at | 1289 | 2.691 | 0.4047 | Yes | ||
| 16 | LIG1 | 1416641_at | 1339 | 2.636 | 0.4221 | Yes | ||
| 17 | CDKN1A | 1421679_a_at 1424638_at | 1464 | 2.524 | 0.4353 | Yes | ||
| 18 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 1662 | 2.360 | 0.4438 | Yes | ||
| 19 | PCNA | 1417947_at | 2013 | 2.123 | 0.4437 | Yes | ||
| 20 | POLE2 | 1427094_at 1431440_at | 2258 | 1.996 | 0.4474 | Yes | ||
| 21 | PSMA7 | 1423567_a_at 1423568_at | 2445 | 1.921 | 0.4532 | Yes | ||
| 22 | RFC4 | 1424321_at 1438161_s_at | 2456 | 1.914 | 0.4670 | Yes | ||
| 23 | PSME1 | 1417056_at | 2471 | 1.911 | 0.4806 | Yes | ||
| 24 | PSMC5 | 1415740_at | 2802 | 1.787 | 0.4788 | Yes | ||
| 25 | POLA2 | 1425794_at 1448369_at 1456713_at | 2897 | 1.755 | 0.4876 | Yes | ||
| 26 | PSMB9 | 1450696_at | 3175 | 1.672 | 0.4874 | No | ||
| 27 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 3670 | 1.514 | 0.4761 | No | ||
| 28 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 4450 | 1.300 | 0.4502 | No | ||
| 29 | PSMD3 | 1448479_at | 4890 | 1.188 | 0.4389 | No | ||
| 30 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 5339 | 1.097 | 0.4266 | No | ||
| 31 | ORC2L | 1418225_at 1418226_at 1418227_at | 5471 | 1.073 | 0.4286 | No | ||
| 32 | CCND1 | 1417419_at 1417420_at 1448698_at | 5473 | 1.073 | 0.4366 | No | ||
| 33 | PSMD8 | 1423296_at | 5613 | 1.045 | 0.4380 | No | ||
| 34 | CDK2 | 1416873_a_at 1447617_at | 5848 | 1.002 | 0.4348 | No | ||
| 35 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 5987 | 0.974 | 0.4357 | No | ||
| 36 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 6010 | 0.970 | 0.4419 | No | ||
| 37 | CUL1 | 1448174_at 1459516_at | 6410 | 0.903 | 0.4304 | No | ||
| 38 | PSMC3 | 1416282_at | 6712 | 0.847 | 0.4230 | No | ||
| 39 | CDC45L | 1416575_at 1457838_at | 7250 | 0.755 | 0.4040 | No | ||
| 40 | ORC5L | 1415830_at 1441601_at | 7918 | 0.644 | 0.3783 | No | ||
| 41 | WEE1 | 1416773_at 1416774_at | 7995 | 0.632 | 0.3796 | No | ||
| 42 | RPA2 | 1416433_at 1454011_a_at | 8956 | 0.480 | 0.3392 | No | ||
| 43 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 9110 | 0.456 | 0.3356 | No | ||
| 44 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 9310 | 0.427 | 0.3297 | No | ||
| 45 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 9594 | 0.387 | 0.3196 | No | ||
| 46 | PSMB3 | 1417052_at | 10408 | 0.259 | 0.2844 | No | ||
| 47 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 10721 | 0.212 | 0.2717 | No | ||
| 48 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 11482 | 0.097 | 0.2376 | No | ||
| 49 | PSME3 | 1418078_at 1418079_at 1438509_at | 11864 | 0.036 | 0.2205 | No | ||
| 50 | MCM3 | 1420029_at 1426653_at | 11962 | 0.020 | 0.2162 | No | ||
| 51 | PSMD2 | 1415831_at | 12524 | -0.076 | 0.1911 | No | ||
| 52 | PSMB2 | 1444377_at 1448262_at 1459641_at | 13087 | -0.172 | 0.1666 | No | ||
| 53 | POLD1 | 1448187_at 1456055_x_at | 13226 | -0.194 | 0.1618 | No | ||
| 54 | POLD2 | 1448277_at | 13894 | -0.306 | 0.1335 | No | ||
| 55 | PSMB5 | 1415676_a_at | 14248 | -0.371 | 0.1202 | No | ||
| 56 | RFC5 | 1452917_at | 15014 | -0.527 | 0.0891 | No | ||
| 57 | PSMA5 | 1424681_a_at 1434356_a_at | 15216 | -0.567 | 0.0841 | No | ||
| 58 | PSMB6 | 1448822_at | 16305 | -0.813 | 0.0404 | No | ||
| 59 | PSMD10 | 1436559_a_at | 16312 | -0.814 | 0.0462 | No | ||
| 60 | PSMC4 | 1416290_a_at 1416291_at | 16427 | -0.843 | 0.0473 | No | ||
| 61 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 16482 | -0.856 | 0.0512 | No | ||
| 62 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 16488 | -0.857 | 0.0573 | No | ||
| 63 | FEN1 | 1421731_a_at 1436454_x_at | 16603 | -0.886 | 0.0587 | No | ||
| 64 | PSMB4 | 1438984_x_at 1451205_at | 17005 | -0.978 | 0.0477 | No | ||
| 65 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 17107 | -1.001 | 0.0505 | No | ||
| 66 | CCNH | 1418585_at | 17242 | -1.028 | 0.0520 | No | ||
| 67 | ORC1L | 1422663_at 1443172_at | 17521 | -1.096 | 0.0475 | No | ||
| 68 | RPA3 | 1448938_at | 18465 | -1.391 | 0.0147 | No | ||
| 69 | PSMD6 | 1423696_a_at 1423697_at | 18467 | -1.392 | 0.0250 | No | ||
| 70 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 18656 | -1.474 | 0.0274 | No | ||
| 71 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 18968 | -1.611 | 0.0252 | No | ||
| 72 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 19180 | -1.697 | 0.0282 | No | ||
| 73 | MCM5 | 1415945_at 1436808_x_at | 19397 | -1.799 | 0.0317 | No | ||
| 74 | PSMD7 | 1432820_at 1451056_at 1454368_at | 20093 | -2.333 | 0.0173 | No | ||
| 75 | MCM2 | 1434079_s_at 1448777_at | 20165 | -2.392 | 0.0319 | No | ||
| 76 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 20212 | -2.424 | 0.0479 | No | ||
| 77 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21334 | -4.136 | 0.0275 | No |

