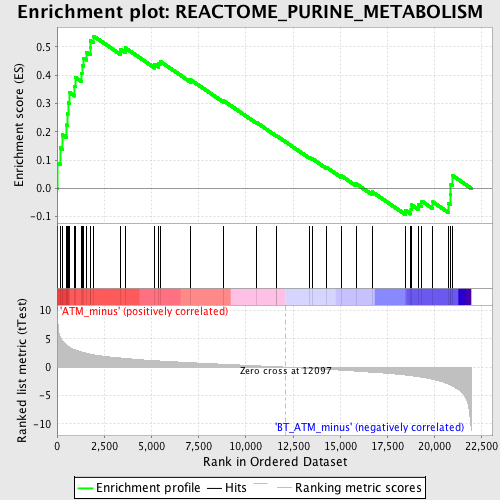
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_PURINE_METABOLISM |
| Enrichment Score (ES) | 0.5377933 |
| Normalized Enrichment Score (NES) | 1.8415331 |
| Nominal p-value | 0.0022371365 |
| FDR q-value | 0.048312485 |
| FWER p-Value | 0.407 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NT5C2 | 1417201_at 1425933_a_at 1442629_at 1443391_at 1448614_at | 38 | 8.230 | 0.0896 | Yes | ||
| 2 | HPRT1 | 1448736_a_at | 161 | 5.328 | 0.1431 | Yes | ||
| 3 | AMPD3 | 1422573_at | 275 | 4.676 | 0.1898 | Yes | ||
| 4 | ADSL | 1418372_at | 485 | 3.944 | 0.2240 | Yes | ||
| 5 | APRT | 1423801_a_at | 538 | 3.812 | 0.2639 | Yes | ||
| 6 | ATP5J | 1416143_at | 578 | 3.701 | 0.3032 | Yes | ||
| 7 | ATP5G1 | 1416020_a_at 1444874_at | 666 | 3.489 | 0.3379 | Yes | ||
| 8 | GUK1 | 1416395_at | 922 | 3.063 | 0.3602 | Yes | ||
| 9 | ATP5E | 1416567_s_at | 962 | 3.031 | 0.3920 | Yes | ||
| 10 | PPAT | 1428543_at 1441770_at 1452831_s_at | 1290 | 2.688 | 0.4069 | Yes | ||
| 11 | ATP5F1 | 1426742_at 1433562_s_at | 1320 | 2.651 | 0.4350 | Yes | ||
| 12 | NME2 | 1448808_a_at | 1415 | 2.576 | 0.4593 | Yes | ||
| 13 | PAICS | 1423564_a_at 1423565_at 1436298_x_at | 1548 | 2.452 | 0.4805 | Yes | ||
| 14 | ADSS | 1460726_at | 1750 | 2.297 | 0.4968 | Yes | ||
| 15 | ATIC | 1428506_at 1452811_at | 1768 | 2.285 | 0.5213 | Yes | ||
| 16 | AMPD2 | 1426757_at 1438941_x_at | 1935 | 2.169 | 0.5378 | Yes | ||
| 17 | NP | 1416530_a_at | 3345 | 1.622 | 0.4914 | No | ||
| 18 | GART | 1416283_at 1424435_a_at 1424436_at 1441911_x_at | 3602 | 1.534 | 0.4967 | No | ||
| 19 | PFAS | 1455496_at | 5172 | 1.130 | 0.4376 | No | ||
| 20 | ATP5O | 1416278_a_at | 5387 | 1.090 | 0.4399 | No | ||
| 21 | IMPDH2 | 1415851_a_at 1415852_at | 5455 | 1.076 | 0.4488 | No | ||
| 22 | ATP5J2 | 1416269_at 1435395_s_at | 7067 | 0.784 | 0.3839 | No | ||
| 23 | ATP5L | 1448203_at | 8810 | 0.503 | 0.3099 | No | ||
| 24 | AK1 | 1422184_a_at 1423988_at | 10545 | 0.238 | 0.2334 | No | ||
| 25 | ATP5I | 1422525_at 1434053_x_at 1450640_x_at | 11608 | 0.078 | 0.1857 | No | ||
| 26 | ATP5C1 | 1416058_s_at 1438809_at | 11634 | 0.072 | 0.1854 | No | ||
| 27 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 13362 | -0.216 | 0.1089 | No | ||
| 28 | NME1 | 1424110_a_at 1435277_x_at | 13512 | -0.242 | 0.1048 | No | ||
| 29 | GMPS | 1433567_at 1435656_at 1455057_at | 14268 | -0.375 | 0.0744 | No | ||
| 30 | ATP5D | 1423716_s_at 1456580_s_at | 15035 | -0.531 | 0.0453 | No | ||
| 31 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 15830 | -0.697 | 0.0168 | No | ||
| 32 | TXNRD1 | 1421529_a_at 1424486_a_at 1424487_x_at | 16680 | -0.909 | -0.0119 | No | ||
| 33 | ADSSL1 | 1449383_at | 18438 | -1.382 | -0.0768 | No | ||
| 34 | ATP5B | 1416829_at | 18738 | -1.511 | -0.0737 | No | ||
| 35 | XDH | 1451006_at | 18758 | -1.518 | -0.0578 | No | ||
| 36 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 19118 | -1.672 | -0.0556 | No | ||
| 37 | RRM1 | 1415878_at 1440073_at 1448127_at | 19304 | -1.758 | -0.0446 | No | ||
| 38 | AMPD1 | 1434722_at | 19901 | -2.167 | -0.0478 | No | ||
| 39 | ATP5H | 1423676_at 1435112_a_at | 20724 | -2.996 | -0.0521 | No | ||
| 40 | DGUOK | 1425228_a_at | 20817 | -3.127 | -0.0216 | No | ||
| 41 | ADA | 1417976_at | 20854 | -3.202 | 0.0123 | No | ||
| 42 | IMPDH1 | 1423239_at | 20928 | -3.336 | 0.0460 | No |

