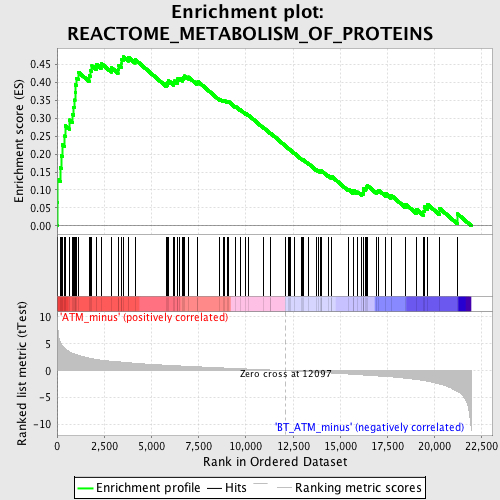
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_METABOLISM_OF_PROTEINS |
| Enrichment Score (ES) | 0.4716477 |
| Normalized Enrichment Score (NES) | 1.8743098 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.036373634 |
| FWER p-Value | 0.278 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DPM1 | 1419353_at 1449438_at | 10 | 9.105 | 0.0659 | Yes | ||
| 2 | EIF2S2 | 1417712_at 1417713_at 1441023_at 1448819_at 1448820_a_at 1456617_a_at | 21 | 8.894 | 0.1303 | Yes | ||
| 3 | EIF2B1 | 1460730_at | 165 | 5.299 | 0.1624 | Yes | ||
| 4 | CCT5 | 1417258_at | 223 | 4.912 | 0.1956 | Yes | ||
| 5 | CCT6A | 1423517_at 1455988_a_at | 288 | 4.587 | 0.2261 | Yes | ||
| 6 | PIGP | 1436038_a_at | 410 | 4.143 | 0.2508 | Yes | ||
| 7 | CCT8 | 1415785_a_at 1436973_at | 437 | 4.047 | 0.2791 | Yes | ||
| 8 | F2 | 1418897_at | 644 | 3.541 | 0.2955 | Yes | ||
| 9 | EEF1D | 1428135_a_at 1439439_x_at 1449506_a_at 1459754_x_at | 818 | 3.213 | 0.3110 | Yes | ||
| 10 | CCT2 | 1416037_a_at 1433534_a_at 1433535_x_at | 876 | 3.134 | 0.3312 | Yes | ||
| 11 | RPL4 | 1425183_a_at | 927 | 3.062 | 0.3513 | Yes | ||
| 12 | RPS3 | 1435151_a_at 1447563_at 1455600_at | 949 | 3.041 | 0.3725 | Yes | ||
| 13 | RPS3A | 1422475_a_at | 969 | 3.020 | 0.3936 | Yes | ||
| 14 | VBP1 | 1421750_a_at | 1038 | 2.958 | 0.4121 | Yes | ||
| 15 | PFDN1 | 1417026_at | 1136 | 2.854 | 0.4285 | Yes | ||
| 16 | TBCA | 1417652_a_at 1437907_a_at 1456205_x_at | 1697 | 2.338 | 0.4199 | Yes | ||
| 17 | EIF4A1 | 1427058_at 1430980_a_at 1434985_a_at | 1752 | 2.297 | 0.4342 | Yes | ||
| 18 | EIF4B | 1426378_at 1426379_at 1426380_at | 1843 | 2.238 | 0.4463 | Yes | ||
| 19 | RPL30 | 1438076_at 1456266_at | 2069 | 2.091 | 0.4513 | Yes | ||
| 20 | PFDN6 | 1415744_at | 2331 | 1.965 | 0.4537 | Yes | ||
| 21 | GSPT2 | 1418109_at 1455721_at | 2868 | 1.763 | 0.4420 | Yes | ||
| 22 | DPM2 | 1415675_at | 3227 | 1.654 | 0.4377 | Yes | ||
| 23 | RPL6 | 1416546_a_at | 3253 | 1.648 | 0.4485 | Yes | ||
| 24 | EEF1G | 1417364_at 1458955_at | 3400 | 1.600 | 0.4535 | Yes | ||
| 25 | EIF4EBP1 | 1417562_at 1417563_at 1434976_x_at | 3405 | 1.597 | 0.4650 | Yes | ||
| 26 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 3509 | 1.562 | 0.4716 | Yes | ||
| 27 | PROS1 | 1426246_at | 3768 | 1.490 | 0.4707 | No | ||
| 28 | PIGF | 1448896_at | 4126 | 1.386 | 0.4645 | No | ||
| 29 | GGCX | 1423554_at 1445898_at | 5778 | 1.013 | 0.3963 | No | ||
| 30 | RPL5 | 1423665_a_at 1447460_at | 5864 | 0.998 | 0.3996 | No | ||
| 31 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 5912 | 0.989 | 0.4047 | No | ||
| 32 | RPL41 | 1422623_x_at 1455578_x_at | 6185 | 0.944 | 0.3991 | No | ||
| 33 | RPL17 | 1423855_x_at 1435791_x_at 1453752_at | 6219 | 0.939 | 0.4045 | No | ||
| 34 | PIGA | 1427305_at 1453497_a_at | 6370 | 0.912 | 0.4042 | No | ||
| 35 | PROZ | 1431721_a_at | 6388 | 0.908 | 0.4101 | No | ||
| 36 | RPS23 | 1460175_at | 6500 | 0.884 | 0.4114 | No | ||
| 37 | RPS16 | 1416404_s_at 1437196_x_at 1455835_at | 6664 | 0.855 | 0.4102 | No | ||
| 38 | EIF5 | 1415723_at 1433631_at 1454663_at 1454664_a_at 1456256_at | 6695 | 0.850 | 0.4150 | No | ||
| 39 | RPS4X | 1416276_a_at | 6757 | 0.839 | 0.4184 | No | ||
| 40 | F7 | 1419321_at | 6950 | 0.808 | 0.4155 | No | ||
| 41 | RPS17 | 1437510_x_at 1438501_at 1438986_x_at 1455662_x_at 1457940_at 1459986_a_at | 7411 | 0.729 | 0.3997 | No | ||
| 42 | RPS24 | 1436064_x_at 1436500_at 1453362_x_at 1455195_at 1456628_x_at | 7459 | 0.721 | 0.4028 | No | ||
| 43 | DHPS | 1434003_a_at 1434004_at | 8624 | 0.532 | 0.3534 | No | ||
| 44 | GPAA1 | 1438152_at 1448225_at 1455954_x_at | 8794 | 0.506 | 0.3494 | No | ||
| 45 | RPL11 | 1417615_a_at | 8851 | 0.495 | 0.3504 | No | ||
| 46 | RPL8 | 1417762_a_at | 9001 | 0.474 | 0.3471 | No | ||
| 47 | CCT3 | 1416024_x_at 1426067_x_at 1439679_at 1448178_a_at 1449645_s_at 1451915_at 1459987_s_at | 9057 | 0.464 | 0.3479 | No | ||
| 48 | RPL37 | 1453729_a_at | 9471 | 0.404 | 0.3320 | No | ||
| 49 | RPS19 | 1449243_a_at 1460442_at | 9687 | 0.373 | 0.3248 | No | ||
| 50 | RPS21 | 1430288_x_at 1433549_x_at 1433721_x_at 1434358_x_at | 9993 | 0.323 | 0.3132 | No | ||
| 51 | RPL37A | 1416217_a_at 1460543_x_at | 10133 | 0.303 | 0.3091 | No | ||
| 52 | RPL18A | 1428152_a_at | 10934 | 0.180 | 0.2738 | No | ||
| 53 | EIF5B | 1434604_at 1434605_at 1435592_at | 11286 | 0.126 | 0.2586 | No | ||
| 54 | PROC | 1418845_at 1444979_at | 11317 | 0.121 | 0.2581 | No | ||
| 55 | EIF4E | 1450909_at 1457489_at 1459371_at | 12117 | -0.004 | 0.2216 | No | ||
| 56 | RPS5 | 1416054_at | 12261 | -0.025 | 0.2152 | No | ||
| 57 | RPS15 | 1416088_a_at 1416089_at | 12283 | -0.031 | 0.2145 | No | ||
| 58 | GAS6 | 1417399_at 1459585_at | 12376 | -0.047 | 0.2106 | No | ||
| 59 | RPS2 | 1431766_x_at | 12559 | -0.084 | 0.2029 | No | ||
| 60 | EIF2B2 | 1434538_x_at 1437428_x_at 1451136_a_at | 12966 | -0.150 | 0.1854 | No | ||
| 61 | RPL28 | 1416074_a_at | 13008 | -0.158 | 0.1846 | No | ||
| 62 | RPL35A | 1417317_s_at | 13040 | -0.165 | 0.1844 | No | ||
| 63 | PLAUR | 1452521_a_at | 13317 | -0.210 | 0.1733 | No | ||
| 64 | PIGH | 1430589_at 1455860_at | 13731 | -0.282 | 0.1565 | No | ||
| 65 | F9 | 1427393_at | 13833 | -0.297 | 0.1540 | No | ||
| 66 | RPS11 | 1424000_a_at | 13960 | -0.319 | 0.1506 | No | ||
| 67 | RPSA | 1416196_at 1448245_at | 13992 | -0.324 | 0.1515 | No | ||
| 68 | RPL19 | 1416219_at | 14005 | -0.326 | 0.1533 | No | ||
| 69 | RPS20 | 1456373_x_at 1456436_x_at | 14354 | -0.389 | 0.1402 | No | ||
| 70 | RPS14 | 1436586_x_at | 14510 | -0.422 | 0.1362 | No | ||
| 71 | RPLP1 | 1416277_a_at | 14533 | -0.426 | 0.1383 | No | ||
| 72 | RPLP2 | 1415879_a_at | 15412 | -0.605 | 0.1025 | No | ||
| 73 | PIGG | 1444640_at | 15676 | -0.667 | 0.0953 | No | ||
| 74 | PIGN | 1421284_at 1425813_at 1425997_a_at 1432115_a_at | 15700 | -0.672 | 0.0992 | No | ||
| 75 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 15919 | -0.719 | 0.0945 | No | ||
| 76 | RPL18 | 1437005_a_at 1450372_a_at | 16132 | -0.771 | 0.0904 | No | ||
| 77 | RPL14 | 1422128_at 1426793_a_at 1433688_x_at 1436688_x_at 1438507_x_at 1438626_x_at | 16207 | -0.788 | 0.0927 | No | ||
| 78 | RPL27A | 1437729_at | 16217 | -0.791 | 0.0981 | No | ||
| 79 | FAU | 1417452_a_at | 16229 | -0.793 | 0.1034 | No | ||
| 80 | SEMA6D | 1446536_at 1448060_at 1453055_at 1459318_at | 16343 | -0.824 | 0.1042 | No | ||
| 81 | EIF2S1 | 1440272_at 1452662_a_at | 16379 | -0.831 | 0.1086 | No | ||
| 82 | TBCC | 1424644_at 1451423_at | 16416 | -0.840 | 0.1131 | No | ||
| 83 | F10 | 1418992_at 1418993_s_at 1449305_at | 16919 | -0.960 | 0.0971 | No | ||
| 84 | RPS15A | 1453467_s_at 1457726_at | 17034 | -0.985 | 0.0991 | No | ||
| 85 | TBCE | 1428282_at 1445668_at | 17406 | -1.064 | 0.0899 | No | ||
| 86 | RPS28 | 1451101_a_at | 17683 | -1.135 | 0.0855 | No | ||
| 87 | RPL39 | 1423032_at 1450840_a_at | 18445 | -1.383 | 0.0608 | No | ||
| 88 | EIF2B5 | 1433886_at 1445908_at | 19021 | -1.627 | 0.0463 | No | ||
| 89 | CCT4 | 1415867_at 1415868_at 1430034_at 1433447_x_at 1438560_x_at 1442569_at 1456082_x_at 1456572_x_at | 19424 | -1.809 | 0.0411 | No | ||
| 90 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 19459 | -1.836 | 0.0529 | No | ||
| 91 | FURIN | 1418518_at | 19626 | -1.944 | 0.0595 | No | ||
| 92 | RPS25 | 1430978_at 1451068_s_at | 20265 | -2.459 | 0.0482 | No | ||
| 93 | EEF2 | 1424736_at 1440784_at 1441414_at | 21207 | -3.861 | 0.0333 | No |

