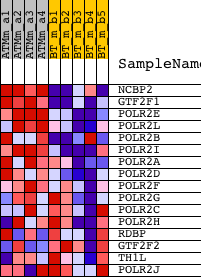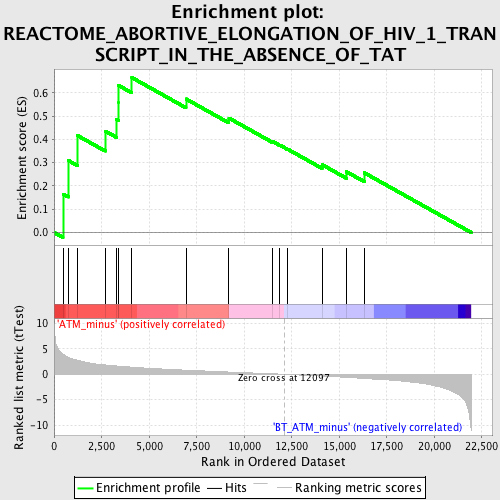
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_ABORTIVE_ELONGATION_OF_HIV_1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT |
| Enrichment Score (ES) | 0.66526777 |
| Normalized Enrichment Score (NES) | 1.7946477 |
| Nominal p-value | 0.0040983604 |
| FDR q-value | 0.061850652 |
| FWER p-Value | 0.62 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCBP2 | 1423045_at 1423046_s_at 1450847_at | 468 | 3.976 | 0.1652 | Yes | ||
| 2 | GTF2F1 | 1417698_at 1417699_at | 749 | 3.340 | 0.3091 | Yes | ||
| 3 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 1231 | 2.751 | 0.4162 | Yes | ||
| 4 | POLR2L | 1428296_at | 2685 | 1.829 | 0.4357 | Yes | ||
| 5 | POLR2B | 1433552_a_at | 3286 | 1.639 | 0.4852 | Yes | ||
| 6 | POLR2I | 1428494_a_at | 3373 | 1.611 | 0.5569 | Yes | ||
| 7 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 3378 | 1.608 | 0.6321 | Yes | ||
| 8 | POLR2D | 1424258_at | 4090 | 1.398 | 0.6653 | Yes | ||
| 9 | POLR2F | 1415754_at 1427402_at | 6937 | 0.810 | 0.5734 | No | ||
| 10 | POLR2G | 1416270_at | 9196 | 0.444 | 0.4912 | No | ||
| 11 | POLR2C | 1416341_at | 11509 | 0.093 | 0.3901 | No | ||
| 12 | POLR2H | 1424473_at | 11875 | 0.034 | 0.3751 | No | ||
| 13 | RDBP | 1450705_at | 12271 | -0.028 | 0.3583 | No | ||
| 14 | GTF2F2 | 1426626_at 1443233_at | 14094 | -0.342 | 0.2913 | No | ||
| 15 | TH1L | 1416198_at | 15387 | -0.599 | 0.2604 | No | ||
| 16 | POLR2J | 1417720_at 1440065_at | 16322 | -0.815 | 0.2560 | No |