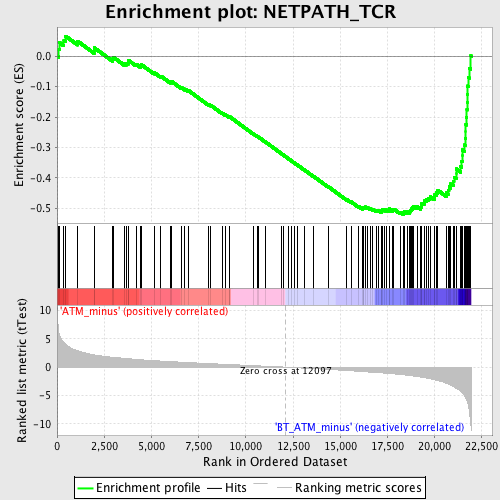
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.5204109 |
| Normalized Enrichment Score (NES) | -2.0455773 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.01121174 |
| FWER p-Value | 0.024 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UNC119 | 1418123_at | 92 | 6.072 | 0.0221 | No | ||
| 2 | CEBPB | 1418901_at 1427844_a_at | 129 | 5.556 | 0.0445 | No | ||
| 3 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 361 | 4.302 | 0.0526 | No | ||
| 4 | SHB | 1434153_at | 450 | 4.012 | 0.0659 | No | ||
| 5 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1090 | 2.905 | 0.0492 | No | ||
| 6 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 1960 | 2.152 | 0.0187 | No | ||
| 7 | PIK3R2 | 1418463_at | 1970 | 2.146 | 0.0276 | No | ||
| 8 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 2937 | 1.741 | -0.0091 | No | ||
| 9 | PLCG1 | 1435149_at 1450360_at | 2985 | 1.724 | -0.0038 | No | ||
| 10 | GRB2 | 1418508_a_at 1449111_a_at | 3588 | 1.538 | -0.0247 | No | ||
| 11 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 3687 | 1.511 | -0.0227 | No | ||
| 12 | PTPN12 | 1422045_a_at 1439705_at 1450478_a_at 1450479_x_at 1455105_at 1459459_at | 3759 | 1.493 | -0.0194 | No | ||
| 13 | NFAM1 | 1425714_a_at 1428790_at | 3791 | 1.482 | -0.0144 | No | ||
| 14 | RAP1A | 1424139_at | 4179 | 1.374 | -0.0262 | No | ||
| 15 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 4425 | 1.307 | -0.0318 | No | ||
| 16 | RASA1 | 1426476_at 1426477_at 1426478_at 1438998_at | 4454 | 1.298 | -0.0274 | No | ||
| 17 | KHDRBS1 | 1418628_at 1418630_at | 5139 | 1.134 | -0.0539 | No | ||
| 18 | ARHGEF6 | 1429012_at 1442292_at | 5496 | 1.067 | -0.0656 | No | ||
| 19 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 5999 | 0.972 | -0.0843 | No | ||
| 20 | PTPN22 | 1417995_at 1442820_at | 6062 | 0.962 | -0.0830 | No | ||
| 21 | MAP2K1 | 1416351_at | 6572 | 0.873 | -0.1026 | No | ||
| 22 | RIPK2 | 1421236_at 1450173_at | 6767 | 0.837 | -0.1078 | No | ||
| 23 | PTPRJ | 1425587_a_at 1425588_at 1427602_at 1427629_at 1443683_at 1455030_at | 6931 | 0.812 | -0.1118 | No | ||
| 24 | SHC1 | 1422853_at 1422854_at | 8026 | 0.627 | -0.1592 | No | ||
| 25 | STK39 | 1419550_a_at 1419551_s_at 1431434_at 1445431_at | 8130 | 0.611 | -0.1613 | No | ||
| 26 | DUSP3 | 1425608_at 1434472_at 1456769_at | 8746 | 0.515 | -0.1872 | No | ||
| 27 | ABI1 | 1423177_a_at 1423178_at 1450890_a_at | 8911 | 0.485 | -0.1926 | No | ||
| 28 | SKAP1 | 1437249_at 1440326_at 1440782_at 1441322_at 1441418_at 1441589_at 1456678_at | 9115 | 0.455 | -0.1999 | No | ||
| 29 | SLA2 | 1437504_at 1451492_at | 9126 | 0.454 | -0.1984 | No | ||
| 30 | DNM2 | 1423629_at 1425135_a_at 1425136_x_at 1432004_a_at 1432005_at 1437938_x_at 1443150_at 1451057_x_at | 10418 | 0.257 | -0.2565 | No | ||
| 31 | ACP1 | 1422716_a_at 1422717_at 1450720_at 1450721_at | 10609 | 0.230 | -0.2642 | No | ||
| 32 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 10669 | 0.219 | -0.2659 | No | ||
| 33 | PTPN11 | 1421196_at 1427699_a_at 1451225_at | 11042 | 0.164 | -0.2823 | No | ||
| 34 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 11878 | 0.033 | -0.3204 | No | ||
| 35 | JAK3 | 1425750_a_at 1446515_at | 11975 | 0.019 | -0.3247 | No | ||
| 36 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 12265 | -0.026 | -0.3378 | No | ||
| 37 | LIME1 | 1416869_x_at 1437589_x_at 1448500_a_at | 12424 | -0.056 | -0.3448 | No | ||
| 38 | FYB | 1452117_a_at | 12591 | -0.090 | -0.3520 | No | ||
| 39 | LAT | 1460651_at | 12741 | -0.112 | -0.3584 | No | ||
| 40 | GAB2 | 1419829_a_at 1420785_at 1439786_at 1446860_at | 13078 | -0.170 | -0.3730 | No | ||
| 41 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 13566 | -0.252 | -0.3943 | No | ||
| 42 | ENAH | 1421624_a_at 1424800_at 1424801_at 1431162_a_at 1442223_at 1446426_at 1458876_at | 14375 | -0.395 | -0.4296 | No | ||
| 43 | WAS | 1419631_at | 15348 | -0.593 | -0.4715 | No | ||
| 44 | SOS1 | 1421884_at 1421885_at 1421886_at | 15572 | -0.643 | -0.4790 | No | ||
| 45 | PTPRC | 1422124_a_at 1440165_at | 15964 | -0.730 | -0.4937 | No | ||
| 46 | FOS | 1423100_at | 16185 | -0.784 | -0.5004 | No | ||
| 47 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 16226 | -0.792 | -0.4988 | No | ||
| 48 | SOS2 | 1452281_at | 16240 | -0.796 | -0.4959 | No | ||
| 49 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 16308 | -0.813 | -0.4955 | No | ||
| 50 | CD2AP | 1420906_at 1420907_at 1420908_at 1460140_at | 16434 | -0.844 | -0.4976 | No | ||
| 51 | DOCK2 | 1422808_s_at 1437282_at 1438334_at 1459382_at | 16591 | -0.881 | -0.5009 | No | ||
| 52 | CREB1 | 1421582_a_at 1421583_at 1423402_at 1428755_at 1452529_a_at 1452901_at | 16714 | -0.917 | -0.5025 | No | ||
| 53 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 16890 | -0.955 | -0.5064 | No | ||
| 54 | PTPN3 | 1443162_at 1455450_at | 16998 | -0.977 | -0.5071 | No | ||
| 55 | ITK | 1417171_at 1430833_at 1452518_a_at 1456836_at 1457120_at | 17183 | -1.015 | -0.5111 | No | ||
| 56 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 17198 | -1.018 | -0.5073 | No | ||
| 57 | NEDD9 | 1422818_at 1437132_x_at 1447885_x_at 1450767_at | 17215 | -1.022 | -0.5036 | No | ||
| 58 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 17336 | -1.044 | -0.5046 | No | ||
| 59 | CD8A | 1440164_x_at 1440811_x_at | 17457 | -1.076 | -0.5054 | No | ||
| 60 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 17587 | -1.112 | -0.5065 | No | ||
| 61 | PRKCQ | 1426044_a_at | 17590 | -1.113 | -0.5018 | No | ||
| 62 | FYN | 1417558_at 1441647_at 1448765_at | 17777 | -1.164 | -0.5053 | No | ||
| 63 | GRAP2 | 1456432_at | 17838 | -1.184 | -0.5029 | No | ||
| 64 | SKAP2 | 1418895_at 1457097_at 1460623_at | 18171 | -1.290 | -0.5125 | No | ||
| 65 | JUN | 1417409_at 1448694_at | 18344 | -1.347 | -0.5146 | Yes | ||
| 66 | SRC | 1423240_at 1450918_s_at | 18385 | -1.363 | -0.5105 | Yes | ||
| 67 | ARHGEF7 | 1424482_at 1449066_a_at | 18531 | -1.417 | -0.5110 | Yes | ||
| 68 | TRAT1 | 1427532_at 1437561_at | 18648 | -1.470 | -0.5100 | Yes | ||
| 69 | DBNL | 1460334_at | 18699 | -1.495 | -0.5058 | Yes | ||
| 70 | PTPN6 | 1456694_x_at 1460188_at | 18763 | -1.521 | -0.5021 | Yes | ||
| 71 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 18827 | -1.548 | -0.4983 | Yes | ||
| 72 | DTX1 | 1425822_a_at 1458643_at | 18881 | -1.573 | -0.4939 | Yes | ||
| 73 | PRKD2 | 1434333_a_at 1434334_at 1436589_x_at 1437509_x_at 1456075_at | 19064 | -1.646 | -0.4951 | Yes | ||
| 74 | CARD11 | 1430257_at 1435996_at | 19248 | -1.735 | -0.4959 | Yes | ||
| 75 | CD247 | 1420716_at 1426079_at 1426396_at 1428059_at 1428060_at 1438392_at 1452539_a_at | 19297 | -1.756 | -0.4905 | Yes | ||
| 76 | EVL | 1434920_a_at 1440885_at 1445957_at 1450106_a_at 1456453_at 1458889_at | 19315 | -1.764 | -0.4837 | Yes | ||
| 77 | HOMER3 | 1424859_at | 19437 | -1.818 | -0.4813 | Yes | ||
| 78 | SH2D2A | 1449105_at | 19475 | -1.842 | -0.4751 | Yes | ||
| 79 | SLA | 1420818_at 1420819_at 1441761_at 1447813_x_at | 19556 | -1.889 | -0.4705 | Yes | ||
| 80 | LCP2 | 1418641_at 1418642_at | 19655 | -1.963 | -0.4665 | Yes | ||
| 81 | MAPK1 | 1419568_at 1426585_s_at 1442876_at 1453104_at | 19751 | -2.036 | -0.4621 | Yes | ||
| 82 | PAG1 | 1423002_at 1440033_at 1443319_at 1456403_at | 19979 | -2.231 | -0.4628 | Yes | ||
| 83 | CD3G | 1419178_at | 20005 | -2.256 | -0.4542 | Yes | ||
| 84 | RAPGEF1 | 1421146_at 1427006_at 1441386_at | 20099 | -2.341 | -0.4483 | Yes | ||
| 85 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 20162 | -2.391 | -0.4408 | Yes | ||
| 86 | TUBB2A | 1427838_at | 20618 | -2.851 | -0.4493 | Yes | ||
| 87 | CBLB | 1437304_at 1455082_at 1458469_at | 20711 | -2.984 | -0.4406 | Yes | ||
| 88 | CD3D | 1422828_at | 20755 | -3.039 | -0.4294 | Yes | ||
| 89 | STAT5A | 1421469_a_at 1450259_a_at | 20852 | -3.200 | -0.4199 | Yes | ||
| 90 | YWHAQ | 1420828_s_at 1420829_a_at 1420830_x_at 1432842_s_at 1437608_x_at 1454378_at 1460590_s_at 1460621_x_at | 20980 | -3.433 | -0.4108 | Yes | ||
| 91 | VASP | 1451097_at | 21020 | -3.497 | -0.3975 | Yes | ||
| 92 | SH2D3C | 1415886_at | 21136 | -3.731 | -0.3866 | Yes | ||
| 93 | ARHGDIB | 1426454_at | 21139 | -3.736 | -0.3705 | Yes | ||
| 94 | CISH | 1448724_at | 21375 | -4.255 | -0.3628 | Yes | ||
| 95 | CD3E | 1422105_at 1445748_at | 21430 | -4.440 | -0.3461 | Yes | ||
| 96 | CD4 | 1419696_at 1427779_a_at | 21452 | -4.512 | -0.3275 | Yes | ||
| 97 | SH3BP2 | 1448328_at | 21456 | -4.518 | -0.3081 | Yes | ||
| 98 | CD5 | 1418353_at | 21580 | -5.050 | -0.2918 | Yes | ||
| 99 | LAX1 | 1438687_at | 21610 | -5.229 | -0.2705 | Yes | ||
| 100 | VAV1 | 1422932_a_at | 21645 | -5.494 | -0.2483 | Yes | ||
| 101 | WASF2 | 1438683_at | 21647 | -5.497 | -0.2245 | Yes | ||
| 102 | DEF6 | 1452796_at | 21691 | -5.764 | -0.2015 | Yes | ||
| 103 | PSTPIP1 | 1424560_at | 21704 | -5.891 | -0.1765 | Yes | ||
| 104 | STAT5B | 1422102_a_at 1422103_a_at | 21715 | -6.071 | -0.1507 | Yes | ||
| 105 | PTK2B | 1434653_at 1442437_at 1442927_at | 21724 | -6.115 | -0.1246 | Yes | ||
| 106 | TXK | 1425785_a_at | 21735 | -6.239 | -0.0980 | Yes | ||
| 107 | GRAP | 1429387_at | 21781 | -6.889 | -0.0702 | Yes | ||
| 108 | SIT1 | 1418751_at | 21824 | -7.604 | -0.0392 | Yes | ||
| 109 | GIT2 | 1423391_at 1432160_at 1435925_at | 21914 | -10.198 | 0.0009 | Yes |

