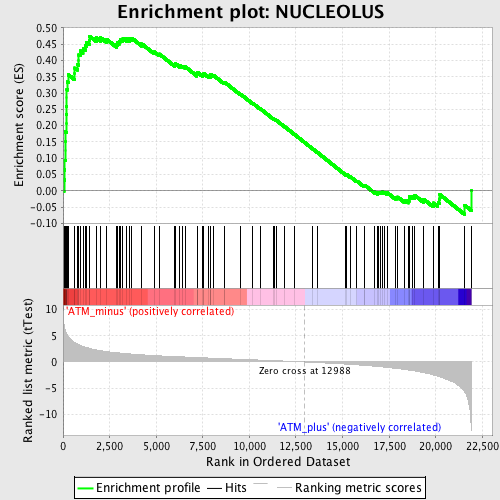
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | NUCLEOLUS |
| Enrichment Score (ES) | 0.47482073 |
| Normalized Enrichment Score (NES) | 1.8819375 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.024603875 |
| FWER p-Value | 0.323 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LYAR | 1417511_at | 49 | 7.195 | 0.0334 | Yes | ||
| 2 | DDX21 | 1448270_at 1448271_a_at | 78 | 6.474 | 0.0643 | Yes | ||
| 3 | NOL5A | 1426533_at 1455035_s_at | 98 | 6.200 | 0.0942 | Yes | ||
| 4 | RPP30 | 1423373_at | 115 | 5.947 | 0.1229 | Yes | ||
| 5 | CIRH1A | 1423884_at | 116 | 5.946 | 0.1524 | Yes | ||
| 6 | MKI67IP | 1424001_at | 122 | 5.908 | 0.1815 | Yes | ||
| 7 | NOLA2 | 1416605_at 1416606_s_at | 167 | 5.483 | 0.2067 | Yes | ||
| 8 | NUFIP1 | 1448802_at | 170 | 5.457 | 0.2337 | Yes | ||
| 9 | UTP18 | 1447266_at 1454817_at | 185 | 5.342 | 0.2595 | Yes | ||
| 10 | MYBBP1A | 1423430_at 1423431_a_at | 201 | 5.249 | 0.2849 | Yes | ||
| 11 | PARP1 | 1422502_at 1422503_s_at 1435368_a_at 1443573_at | 202 | 5.247 | 0.3109 | Yes | ||
| 12 | IKBKAP | 1424142_at 1451254_at | 240 | 5.062 | 0.3343 | Yes | ||
| 13 | RPS19BP1 | 1455174_at | 289 | 4.824 | 0.3561 | Yes | ||
| 14 | MORF4L2 | 1415778_at 1439413_x_at | 627 | 3.693 | 0.3589 | Yes | ||
| 15 | CD3EAP | 1430138_at | 632 | 3.687 | 0.3770 | Yes | ||
| 16 | XRN2 | 1422842_at 1450777_at | 775 | 3.426 | 0.3875 | Yes | ||
| 17 | NOL1 | 1424019_at | 843 | 3.287 | 0.4008 | Yes | ||
| 18 | NHP2L1 | 1416973_at | 846 | 3.280 | 0.4170 | Yes | ||
| 19 | BXDC5 | 1428020_at 1431784_a_at 1459573_at | 916 | 3.164 | 0.4295 | Yes | ||
| 20 | EXOSC9 | 1418462_at | 1068 | 2.979 | 0.4374 | Yes | ||
| 21 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1203 | 2.812 | 0.4452 | Yes | ||
| 22 | SDAD1 | 1437088_at 1452454_at | 1251 | 2.766 | 0.4567 | Yes | ||
| 23 | POLR1E | 1419058_at 1435057_x_at | 1409 | 2.616 | 0.4625 | Yes | ||
| 24 | CDKN2A | 1450140_a_at | 1423 | 2.597 | 0.4748 | Yes | ||
| 25 | TAF5 | 1435474_at | 1781 | 2.291 | 0.4698 | No | ||
| 26 | FGF18 | 1449545_at | 2013 | 2.147 | 0.4699 | No | ||
| 27 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 2345 | 1.975 | 0.4646 | No | ||
| 28 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 2868 | 1.765 | 0.4494 | No | ||
| 29 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 2923 | 1.744 | 0.4556 | No | ||
| 30 | DDX56 | 1423815_at | 3036 | 1.704 | 0.4589 | No | ||
| 31 | ILF2 | 1417948_s_at 1417949_at | 3085 | 1.688 | 0.4651 | No | ||
| 32 | DDX24 | 1415713_a_at 1441631_at | 3213 | 1.649 | 0.4675 | No | ||
| 33 | RPS7 | 1435593_x_at | 3407 | 1.589 | 0.4665 | No | ||
| 34 | ELP3 | 1426643_at 1436762_x_at 1437338_x_at 1443452_at | 3571 | 1.537 | 0.4667 | No | ||
| 35 | POP1 | 1428458_at 1441002_at | 3683 | 1.505 | 0.4690 | No | ||
| 36 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 4229 | 1.366 | 0.4509 | No | ||
| 37 | DDX47 | 1423752_at 1438380_at 1438381_x_at | 4895 | 1.221 | 0.4265 | No | ||
| 38 | DDX54 | 1438853_x_at 1447882_x_at 1460396_at | 5164 | 1.171 | 0.4200 | No | ||
| 39 | TSPYL2 | 1434849_at | 6007 | 1.025 | 0.3865 | No | ||
| 40 | TCOF1 | 1423600_a_at 1423601_s_at 1424643_at | 6025 | 1.022 | 0.3908 | No | ||
| 41 | ASNA1 | 1418292_at | 6275 | 0.986 | 0.3843 | No | ||
| 42 | GNL3 | 1433656_a_at 1440766_at | 6421 | 0.963 | 0.3825 | No | ||
| 43 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 6566 | 0.943 | 0.3806 | No | ||
| 44 | NOL6 | 1424472_at 1425629_a_at | 7189 | 0.845 | 0.3563 | No | ||
| 45 | RPL11 | 1417615_a_at | 7194 | 0.844 | 0.3603 | No | ||
| 46 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 7230 | 0.837 | 0.3628 | No | ||
| 47 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 7499 | 0.796 | 0.3545 | No | ||
| 48 | APTX | 1450921_at | 7520 | 0.793 | 0.3575 | No | ||
| 49 | IMP3 | 1416210_at | 7559 | 0.786 | 0.3597 | No | ||
| 50 | WRN | 1425074_at 1425982_a_at 1450163_a_at | 7800 | 0.754 | 0.3524 | No | ||
| 51 | PNMA1 | 1429224_at | 7907 | 0.737 | 0.3512 | No | ||
| 52 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 7926 | 0.736 | 0.3541 | No | ||
| 53 | NOL4 | 1430189_at 1441447_at 1456387_at | 7928 | 0.736 | 0.3577 | No | ||
| 54 | RPP40 | 1424792_at 1443750_s_at | 8057 | 0.718 | 0.3554 | No | ||
| 55 | SRP68 | 1433708_at | 8656 | 0.625 | 0.3311 | No | ||
| 56 | DDX11 | 1438447_at 1441062_at | 8688 | 0.620 | 0.3327 | No | ||
| 57 | THAP2 | 1421177_at 1455336_at | 9528 | 0.500 | 0.2968 | No | ||
| 58 | EXOSC10 | 1423587_a_at 1431832_x_at 1437294_at 1454021_a_at | 10171 | 0.415 | 0.2695 | No | ||
| 59 | KRR1 | 1420139_s_at 1428084_at 1444726_at 1452658_at | 10572 | 0.356 | 0.2529 | No | ||
| 60 | NEK11 | 1441525_at 1445012_at | 11317 | 0.251 | 0.2201 | No | ||
| 61 | NOL8 | 1428950_s_at 1428951_at 1452974_at | 11360 | 0.246 | 0.2194 | No | ||
| 62 | POP7 | 1449162_at | 11457 | 0.232 | 0.2161 | No | ||
| 63 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 11905 | 0.165 | 0.1965 | No | ||
| 64 | NOLA3 | 1423210_a_at 1423211_at | 12440 | 0.086 | 0.1725 | No | ||
| 65 | SLC29A2 | 1447748_x_at 1448257_at | 13404 | -0.072 | 0.1287 | No | ||
| 66 | MPHOSPH1 | 1419792_at 1439695_a_at 1440924_at 1449612_x_at | 13636 | -0.110 | 0.1187 | No | ||
| 67 | UTP20 | 1452252_at 1459028_at | 15183 | -0.401 | 0.0499 | No | ||
| 68 | PTBP1 | 1424874_a_at 1450443_at 1458284_at | 15242 | -0.414 | 0.0493 | No | ||
| 69 | COIL | 1416692_at | 15424 | -0.462 | 0.0433 | No | ||
| 70 | CCNT1 | 1419313_at 1456477_at | 15780 | -0.550 | 0.0298 | No | ||
| 71 | IMP4 | 1426213_at 1426214_at | 16163 | -0.644 | 0.0155 | No | ||
| 72 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 16190 | -0.655 | 0.0176 | No | ||
| 73 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 16738 | -0.819 | -0.0034 | No | ||
| 74 | MPHOSPH10 | 1429080_at 1431388_at | 16898 | -0.868 | -0.0064 | No | ||
| 75 | NOC4L | 1423826_at 1423827_s_at 1438095_x_at 1443794_x_at | 16956 | -0.886 | -0.0046 | No | ||
| 76 | SURF6 | 1416864_at 1459331_at | 17038 | -0.916 | -0.0038 | No | ||
| 77 | GEMIN4 | 1433622_at | 17135 | -0.946 | -0.0035 | No | ||
| 78 | UBTF | 1453097_a_at 1455490_at 1460304_a_at | 17261 | -0.987 | -0.0043 | No | ||
| 79 | ERCC6 | 1441595_at 1442604_at | 17408 | -1.033 | -0.0059 | No | ||
| 80 | KLHL7 | 1428091_at 1452660_s_at | 17837 | -1.208 | -0.0195 | No | ||
| 81 | NOL3 | 1444786_at 1451503_at | 17964 | -1.263 | -0.0190 | No | ||
| 82 | PML | 1448757_at 1456103_at 1459137_at | 18354 | -1.441 | -0.0296 | No | ||
| 83 | REXO4 | 1434113_a_at 1434114_at | 18526 | -1.527 | -0.0299 | No | ||
| 84 | RPS19 | 1449243_a_at 1460442_at | 18579 | -1.547 | -0.0246 | No | ||
| 85 | RPL35 | 1416243_a_at 1434231_x_at 1436840_x_at 1454833_at 1454856_x_at 1455950_x_at | 18589 | -1.550 | -0.0173 | No | ||
| 86 | SMUG1 | 1428763_at | 18756 | -1.640 | -0.0168 | No | ||
| 87 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 18886 | -1.709 | -0.0142 | No | ||
| 88 | DEDD2 | 1452070_at | 19361 | -2.047 | -0.0258 | No | ||
| 89 | ZNF346 | 1417088_at 1448581_at | 19880 | -2.456 | -0.0373 | No | ||
| 90 | DNAJB9 | 1417191_at | 20131 | -2.729 | -0.0352 | No | ||
| 91 | EMG1 | 1416393_at | 20217 | -2.820 | -0.0251 | No | ||
| 92 | DEDD | 1432000_a_at 1434994_at 1434995_s_at | 20229 | -2.833 | -0.0116 | No | ||
| 93 | NSUN2 | 1423850_at 1453759_at | 21565 | -5.746 | -0.0442 | No | ||
| 94 | EXOSC1 | 1432052_at 1452012_a_at 1453359_at | 21926 | -12.304 | 0.0004 | No |

