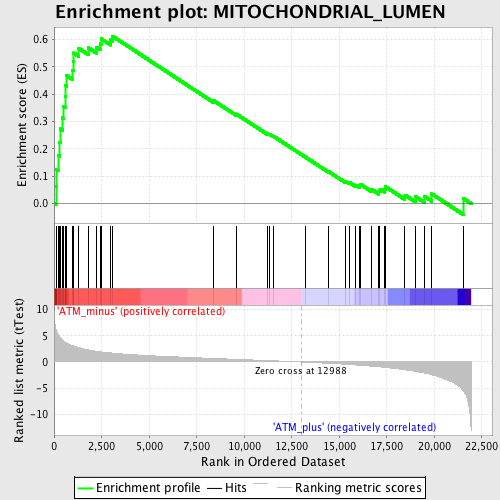
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | MITOCHONDRIAL_LUMEN |
| Enrichment Score (ES) | 0.6125759 |
| Normalized Enrichment Score (NES) | 2.048234 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.007866447 |
| FWER p-Value | 0.033 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ETFA | 1423972_at | 100 | 6.170 | 0.0621 | Yes | ||
| 2 | MRPS22 | 1416595_at 1443032_at | 112 | 5.973 | 0.1260 | Yes | ||
| 3 | PITRM1 | 1426453_at 1456577_x_at | 220 | 5.167 | 0.1769 | Yes | ||
| 4 | MRPL12 | 1452048_at | 298 | 4.780 | 0.2250 | Yes | ||
| 5 | MRPL55 | 1429453_a_at | 336 | 4.608 | 0.2731 | Yes | ||
| 6 | ATP5F1 | 1426742_at 1433562_s_at | 441 | 4.198 | 0.3137 | Yes | ||
| 7 | MRPS36 | 1423242_at | 489 | 4.052 | 0.3553 | Yes | ||
| 8 | MRPS28 | 1452585_at | 583 | 3.786 | 0.3919 | Yes | ||
| 9 | MRPS15 | 1430100_at 1449294_at 1456109_a_at | 600 | 3.752 | 0.4317 | Yes | ||
| 10 | MRPS18C | 1427901_at | 656 | 3.647 | 0.4686 | Yes | ||
| 11 | MRPL51 | 1416877_a_at 1416878_at 1416879_at | 960 | 3.102 | 0.4882 | Yes | ||
| 12 | CS | 1422577_at 1422578_at 1450667_a_at | 1003 | 3.055 | 0.5193 | Yes | ||
| 13 | MRPS35 | 1452111_at | 1026 | 3.019 | 0.5509 | Yes | ||
| 14 | SUPV3L1 | 1432623_at 1460557_at | 1292 | 2.731 | 0.5683 | Yes | ||
| 15 | MRPL40 | 1448849_at | 1796 | 2.281 | 0.5699 | Yes | ||
| 16 | TFB2M | 1423441_at | 2220 | 2.040 | 0.5726 | Yes | ||
| 17 | GRPEL1 | 1417320_at | 2421 | 1.942 | 0.5845 | Yes | ||
| 18 | MRPS12 | 1448799_s_at | 2475 | 1.912 | 0.6027 | Yes | ||
| 19 | ETFB | 1428181_at | 2973 | 1.726 | 0.5986 | Yes | ||
| 20 | NFS1 | 1416373_at 1431431_a_at 1442017_at | 3069 | 1.693 | 0.6126 | Yes | ||
| 21 | BCKDK | 1443813_x_at 1460644_at | 8367 | 0.668 | 0.3778 | No | ||
| 22 | BCKDHA | 1416647_at | 9580 | 0.493 | 0.3278 | No | ||
| 23 | MRPS16 | 1448869_a_at | 11237 | 0.264 | 0.2550 | No | ||
| 24 | MRPS24 | 1438563_s_at 1450865_s_at | 11307 | 0.253 | 0.2546 | No | ||
| 25 | MRPL10 | 1418112_at | 11538 | 0.221 | 0.2465 | No | ||
| 26 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 13197 | -0.035 | 0.1711 | No | ||
| 27 | POLG2 | 1450816_at | 14430 | -0.258 | 0.1176 | No | ||
| 28 | BCKDHB | 1427153_at | 15311 | -0.431 | 0.0821 | No | ||
| 29 | MRPS21 | 1422451_at | 15528 | -0.489 | 0.0775 | No | ||
| 30 | MRPL23 | 1416948_at | 15873 | -0.571 | 0.0680 | No | ||
| 31 | TIMM44 | 1417670_at 1439371_x_at 1448801_a_at | 16051 | -0.616 | 0.0665 | No | ||
| 32 | ACADM | 1415984_at | 16096 | -0.628 | 0.0713 | No | ||
| 33 | MRPL41 | 1428588_a_at 1428589_at 1428590_at | 16703 | -0.810 | 0.0524 | No | ||
| 34 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 17087 | -0.932 | 0.0449 | No | ||
| 35 | DBT | 1440016_at 1449118_at | 17138 | -0.947 | 0.0529 | No | ||
| 36 | MRPL52 | 1415761_at 1415762_x_at 1455366_at 1460701_a_at | 17377 | -1.023 | 0.0530 | No | ||
| 37 | DNAJA3 | 1420629_a_at 1432066_at 1449935_a_at | 17431 | -1.039 | 0.0618 | No | ||
| 38 | MRPS11 | 1455233_at 1460195_at | 18454 | -1.488 | 0.0312 | No | ||
| 39 | MRPS18A | 1416984_at 1457813_at | 19006 | -1.801 | 0.0255 | No | ||
| 40 | CASQ1 | 1422598_at | 19471 | -2.111 | 0.0271 | No | ||
| 41 | ALDH4A1 | 1452375_at | 19866 | -2.439 | 0.0355 | No | ||
| 42 | MRPS10 | 1428153_at 1458999_at | 21513 | -5.460 | 0.0192 | No |

