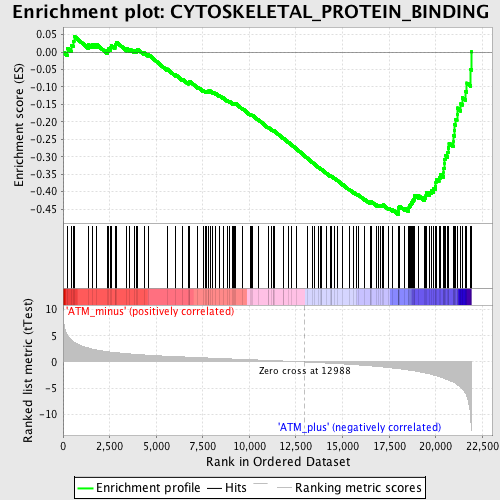
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CYTOSKELETAL_PROTEIN_BINDING |
| Enrichment Score (ES) | -0.4652797 |
| Normalized Enrichment Score (NES) | -1.7569966 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.59245664 |
| FWER p-Value | 0.903 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APOE | 1432466_a_at | 226 | 5.127 | 0.0115 | No | ||
| 2 | KATNA1 | 1450949_at | 451 | 4.176 | 0.0190 | No | ||
| 3 | LRPPRC | 1424353_at | 544 | 3.881 | 0.0314 | No | ||
| 4 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 587 | 3.779 | 0.0455 | No | ||
| 5 | TNNI3 | 1422536_at | 1360 | 2.672 | 0.0215 | No | ||
| 6 | BRCA2 | 1419076_a_at | 1579 | 2.450 | 0.0220 | No | ||
| 7 | UXT | 1418986_a_at | 1806 | 2.274 | 0.0213 | No | ||
| 8 | TNNT1 | 1419606_a_at | 2373 | 1.962 | 0.0037 | No | ||
| 9 | ARL8B | 1418819_at 1428137_at 1428138_s_at | 2439 | 1.931 | 0.0090 | No | ||
| 10 | BIRC5 | 1424278_a_at | 2548 | 1.880 | 0.0120 | No | ||
| 11 | ABLIM1 | 1442376_at 1453103_at 1454708_at 1460120_at | 2572 | 1.869 | 0.0189 | No | ||
| 12 | NDE1 | 1435737_a_at | 2788 | 1.791 | 0.0167 | No | ||
| 13 | MYO9B | 1418031_at 1438533_at 1459110_at | 2831 | 1.775 | 0.0223 | No | ||
| 14 | PARVG | 1416875_at 1416876_at | 2878 | 1.761 | 0.0277 | No | ||
| 15 | TRIM63 | 1438704_at | 3424 | 1.585 | 0.0095 | No | ||
| 16 | HPCA | 1425833_a_at 1450930_at | 3589 | 1.531 | 0.0085 | No | ||
| 17 | ABI2 | 1433985_at 1436984_at 1444783_at 1458053_at 1460153_at | 3825 | 1.468 | 0.0040 | No | ||
| 18 | RABGAP1 | 1424188_at 1441217_at 1441800_at 1443535_at 1457370_at 1460486_at | 3942 | 1.436 | 0.0048 | No | ||
| 19 | ABRA | 1458455_at | 4000 | 1.421 | 0.0082 | No | ||
| 20 | VCL | 1416156_at 1416157_at 1445256_at | 4357 | 1.338 | -0.0024 | No | ||
| 21 | PKD2L1 | 1457105_at | 4562 | 1.286 | -0.0063 | No | ||
| 22 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 5586 | 1.092 | -0.0485 | No | ||
| 23 | SSH1 | 1433874_at 1438253_at 1439237_a_at 1455854_a_at | 6034 | 1.021 | -0.0647 | No | ||
| 24 | TARDBP | 1423723_s_at 1428467_at 1434419_s_at 1436318_at 1455655_a_at | 6403 | 0.966 | -0.0775 | No | ||
| 25 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 6716 | 0.917 | -0.0879 | No | ||
| 26 | SORBS2 | 1437197_at 1441624_at 1446744_at | 6783 | 0.909 | -0.0870 | No | ||
| 27 | ARPC1A | 1416079_a_at | 6792 | 0.907 | -0.0835 | No | ||
| 28 | GC | 1426547_at | 7236 | 0.836 | -0.1003 | No | ||
| 29 | ACTN2 | 1448327_at 1456968_at | 7526 | 0.793 | -0.1101 | No | ||
| 30 | PKD2 | 1417753_at 1441870_s_at | 7653 | 0.773 | -0.1126 | No | ||
| 31 | MYO6 | 1421120_at 1433942_at 1435559_at | 7711 | 0.766 | -0.1120 | No | ||
| 32 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 7824 | 0.750 | -0.1139 | No | ||
| 33 | MYBPC3 | 1418551_at | 7830 | 0.749 | -0.1110 | No | ||
| 34 | RAB11FIP5 | 1427405_s_at 1434314_s_at | 7911 | 0.737 | -0.1115 | No | ||
| 35 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 8048 | 0.719 | -0.1147 | No | ||
| 36 | SORBS3 | 1419329_at | 8202 | 0.693 | -0.1187 | No | ||
| 37 | S100B | 1419383_at 1434342_at | 8405 | 0.663 | -0.1252 | No | ||
| 38 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 8589 | 0.635 | -0.1308 | No | ||
| 39 | NCALD | 1417568_at 1417569_at 1439730_at 1444466_at 1458037_at | 8813 | 0.602 | -0.1385 | No | ||
| 40 | IPP | 1421771_a_at | 8959 | 0.582 | -0.1427 | No | ||
| 41 | FXYD5 | 1418296_at | 9121 | 0.557 | -0.1477 | No | ||
| 42 | LIMA1 | 1422499_at 1425654_a_at 1435726_at 1435727_s_at 1450629_at | 9146 | 0.553 | -0.1464 | No | ||
| 43 | NRAP | 1421253_at | 9215 | 0.544 | -0.1472 | No | ||
| 44 | STMN1 | 1415849_s_at | 9253 | 0.538 | -0.1466 | No | ||
| 45 | SYNPO | 1427045_at 1434089_at | 9653 | 0.482 | -0.1629 | No | ||
| 46 | VAPB | 1423152_at 1436079_s_at 1445370_at 1458501_at | 10066 | 0.426 | -0.1800 | No | ||
| 47 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 10112 | 0.422 | -0.1802 | No | ||
| 48 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 10178 | 0.414 | -0.1814 | No | ||
| 49 | ADD1 | 1420953_at 1420954_a_at 1439125_at 1450054_at | 10491 | 0.369 | -0.1942 | No | ||
| 50 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 11006 | 0.300 | -0.2165 | No | ||
| 51 | TMOD2 | 1430153_at 1431326_a_at 1435060_at 1437167_at 1439223_at 1451301_at | 11025 | 0.297 | -0.2160 | No | ||
| 52 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 11211 | 0.269 | -0.2234 | No | ||
| 53 | NEXN | 1435649_at 1446084_at | 11277 | 0.259 | -0.2253 | No | ||
| 54 | TNNC1 | 1418370_at | 11289 | 0.257 | -0.2247 | No | ||
| 55 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 11333 | 0.250 | -0.2256 | No | ||
| 56 | PDLIM5 | 1429783_at 1438186_at 1442710_at 1446339_at | 11813 | 0.180 | -0.2468 | No | ||
| 57 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 11843 | 0.175 | -0.2474 | No | ||
| 58 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 12128 | 0.131 | -0.2598 | No | ||
| 59 | CAPZA1 | 1439455_x_at 1440380_at | 12277 | 0.108 | -0.2662 | No | ||
| 60 | ANLN | 1433543_at 1439648_at | 12524 | 0.073 | -0.2771 | No | ||
| 61 | PRNP | 1416130_at 1446507_at 1448233_at | 13124 | -0.024 | -0.3045 | No | ||
| 62 | DST | 1421117_at 1421276_a_at 1423626_at 1442395_at 1446943_at 1450119_at 1458075_at 1459098_at 1459356_at | 13399 | -0.070 | -0.3168 | No | ||
| 63 | DYNC1I1 | 1416361_a_at | 13519 | -0.090 | -0.3219 | No | ||
| 64 | ARFGEF1 | 1415711_at 1437104_at 1444175_at 1456917_at | 13721 | -0.130 | -0.3305 | No | ||
| 65 | CAPZB | 1417259_a_at 1424168_a_at 1431668_at 1453960_a_at | 13809 | -0.145 | -0.3339 | No | ||
| 66 | MAP1S | 1454732_at | 13869 | -0.154 | -0.3360 | No | ||
| 67 | RHCG | 1417362_at | 13894 | -0.160 | -0.3364 | No | ||
| 68 | TNNT2 | 1418726_a_at 1424967_x_at 1440424_at | 14148 | -0.204 | -0.3471 | No | ||
| 69 | SHROOM3 | 1422629_s_at 1432377_x_at 1442952_at 1445401_at 1451854_a_at 1453714_a_at 1454211_a_at | 14335 | -0.241 | -0.3546 | No | ||
| 70 | CGN | 1430329_at 1435155_at | 14392 | -0.250 | -0.3561 | No | ||
| 71 | RHAG | 1419014_at | 14402 | -0.252 | -0.3555 | No | ||
| 72 | TTN | 1427445_a_at 1427446_s_at 1431928_at 1444083_at 1444638_at 1446450_at | 14565 | -0.280 | -0.3617 | No | ||
| 73 | ANK1 | 1419421_at 1425677_a_at 1443291_at 1450627_at 1452512_a_at | 14725 | -0.313 | -0.3677 | No | ||
| 74 | RPH3AL | 1431695_at | 15023 | -0.373 | -0.3797 | No | ||
| 75 | CDK5RAP2 | 1443976_at 1447103_at 1452618_at | 15353 | -0.442 | -0.3929 | No | ||
| 76 | CROCC | 1427338_at | 15600 | -0.509 | -0.4020 | No | ||
| 77 | TUBGCP5 | 1426518_at 1439923_at 1439924_x_at 1457282_x_at | 15762 | -0.548 | -0.4071 | No | ||
| 78 | SVIL | 1427662_at 1460694_s_at 1460735_at | 15863 | -0.568 | -0.4092 | No | ||
| 79 | TMOD4 | 1449969_at | 16199 | -0.659 | -0.4218 | No | ||
| 80 | TLN1 | 1436042_at 1448402_at 1457782_at | 16490 | -0.743 | -0.4319 | No | ||
| 81 | MYOZ1 | 1448636_at 1460202_at | 16496 | -0.744 | -0.4290 | No | ||
| 82 | ATG4C | 1434014_at 1441440_at | 16551 | -0.758 | -0.4282 | No | ||
| 83 | ARPC4 | 1423588_at 1423589_at | 16835 | -0.852 | -0.4376 | No | ||
| 84 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 16946 | -0.884 | -0.4389 | No | ||
| 85 | MEFV | 1460283_at | 17033 | -0.914 | -0.4389 | No | ||
| 86 | MAPK8IP1 | 1425679_a_at 1440619_at | 17141 | -0.947 | -0.4398 | No | ||
| 87 | SPTBN4 | 1425116_a_at | 17180 | -0.960 | -0.4374 | No | ||
| 88 | HOOK3 | 1443857_at 1446737_a_at | 17486 | -1.057 | -0.4469 | No | ||
| 89 | BAIAP2 | 1425656_a_at 1435128_at 1451027_at 1451028_at | 17684 | -1.147 | -0.4511 | No | ||
| 90 | TMOD3 | 1423088_at 1423089_at 1438556_a_at 1439626_at 1455708_at 1456913_at | 17995 | -1.273 | -0.4598 | Yes | ||
| 91 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 18010 | -1.279 | -0.4550 | Yes | ||
| 92 | CTNNA1 | 1437275_at 1437807_x_at 1443662_at 1448149_at | 18012 | -1.280 | -0.4496 | Yes | ||
| 93 | CLASP2 | 1424836_a_at 1427328_a_at 1429976_at 1440802_at 1441726_at 1443320_at 1456911_at | 18015 | -1.280 | -0.4443 | Yes | ||
| 94 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 18076 | -1.306 | -0.4414 | Yes | ||
| 95 | CYFIP1 | 1416329_at 1445327_at 1457134_at 1457615_at | 18345 | -1.438 | -0.4476 | Yes | ||
| 96 | MYH10 | 1441057_at 1452740_at | 18550 | -1.535 | -0.4504 | Yes | ||
| 97 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 18571 | -1.542 | -0.4448 | Yes | ||
| 98 | ADD2 | 1421975_a_at 1435287_at 1451914_a_at | 18600 | -1.552 | -0.4394 | Yes | ||
| 99 | KPTN | 1423438_at 1440025_at | 18657 | -1.575 | -0.4353 | Yes | ||
| 100 | KIF1B | 1423994_at 1423995_at 1425270_at 1451200_at 1451642_at 1451762_a_at 1455182_at 1458426_at | 18709 | -1.603 | -0.4308 | Yes | ||
| 101 | COTL1 | 1416001_a_at 1416002_x_at 1425801_x_at 1436236_x_at 1436838_x_at 1437811_x_at | 18769 | -1.652 | -0.4264 | Yes | ||
| 102 | PACSIN3 | 1416995_at 1431534_at 1437834_s_at 1456191_x_at | 18823 | -1.681 | -0.4217 | Yes | ||
| 103 | ESPN | 1423005_a_at 1458260_at | 18873 | -1.703 | -0.4167 | Yes | ||
| 104 | PXN | 1424027_at 1426085_a_at 1456135_s_at | 18885 | -1.708 | -0.4099 | Yes | ||
| 105 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 19092 | -1.854 | -0.4115 | Yes | ||
| 106 | IQGAP2 | 1433885_at 1459894_at | 19401 | -2.070 | -0.4168 | Yes | ||
| 107 | ARL8A | 1424640_at | 19483 | -2.116 | -0.4114 | Yes | ||
| 108 | SYNE1 | 1421545_a_at 1455225_at 1455493_at | 19490 | -2.120 | -0.4027 | Yes | ||
| 109 | POLB | 1425371_at 1434229_a_at | 19664 | -2.251 | -0.4010 | Yes | ||
| 110 | SCIN | 1450276_a_at | 19781 | -2.361 | -0.3963 | Yes | ||
| 111 | SSH2 | 1456153_at | 19900 | -2.478 | -0.3911 | Yes | ||
| 112 | ABI1 | 1423177_a_at 1423178_at 1450890_a_at | 19987 | -2.566 | -0.3841 | Yes | ||
| 113 | SMTN | 1452469_a_at | 20018 | -2.594 | -0.3744 | Yes | ||
| 114 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 20029 | -2.607 | -0.3638 | Yes | ||
| 115 | TUBGCP3 | 1426707_at | 20188 | -2.789 | -0.3591 | Yes | ||
| 116 | ACTA1 | 1427735_a_at | 20245 | -2.852 | -0.3495 | Yes | ||
| 117 | RAE1 | 1429528_at 1444256_at 1459822_at | 20439 | -3.076 | -0.3453 | Yes | ||
| 118 | DBNL | 1460334_at | 20444 | -3.079 | -0.3323 | Yes | ||
| 119 | ACTN4 | 1423449_a_at 1444258_at | 20471 | -3.113 | -0.3203 | Yes | ||
| 120 | UNC84B | 1433832_at 1439238_at | 20478 | -3.124 | -0.3072 | Yes | ||
| 121 | CENPJ | 1423140_at 1423141_at 1437643_at 1450872_s_at | 20521 | -3.189 | -0.2955 | Yes | ||
| 122 | MTSS1 | 1424826_s_at 1434036_at 1440847_at 1443729_at 1446284_at 1451496_at | 20657 | -3.388 | -0.2873 | Yes | ||
| 123 | CXCR4 | 1448710_at | 20678 | -3.415 | -0.2736 | Yes | ||
| 124 | JUP | 1426873_s_at | 20721 | -3.478 | -0.2607 | Yes | ||
| 125 | GABARAP | 1416937_at | 20951 | -3.828 | -0.2549 | Yes | ||
| 126 | MARK4 | 1439051_a_at 1456618_at | 20958 | -3.846 | -0.2388 | Yes | ||
| 127 | CNN2 | 1450981_at | 21010 | -3.935 | -0.2243 | Yes | ||
| 128 | TUBGCP6 | 1435090_at | 21030 | -3.972 | -0.2083 | Yes | ||
| 129 | ATG4D | 1433918_at 1439130_at | 21087 | -4.115 | -0.1933 | Yes | ||
| 130 | PALLD | 1427228_at 1433768_at 1440635_at 1440693_at | 21154 | -4.277 | -0.1781 | Yes | ||
| 131 | CDC42EP3 | 1422642_at 1450700_at | 21164 | -4.289 | -0.1602 | Yes | ||
| 132 | KIF5B | 1418427_at 1418428_at 1418429_at 1418430_at 1418431_at | 21360 | -4.810 | -0.1486 | Yes | ||
| 133 | MYLIP | 1424988_at | 21433 | -5.083 | -0.1303 | Yes | ||
| 134 | FLNA | 1426677_at | 21604 | -5.948 | -0.1127 | Yes | ||
| 135 | WASF2 | 1438683_at | 21686 | -6.500 | -0.0887 | Yes | ||
| 136 | LSP1 | 1417756_a_at | 21894 | -11.171 | -0.0506 | Yes | ||
| 137 | GABARAPL1 | 1416418_at 1416419_s_at 1442572_at | 21925 | -12.275 | 0.0004 | Yes |