Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
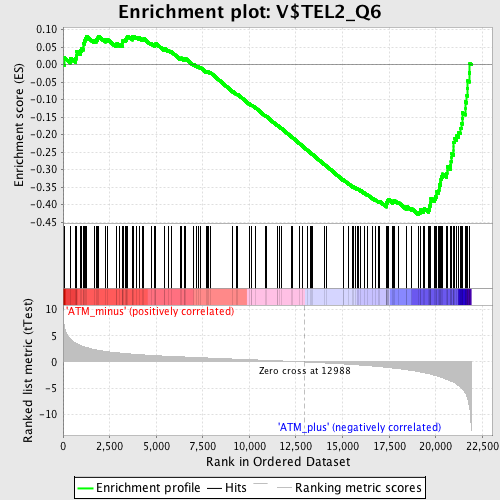
| GeneSet | V$TEL2_Q6 |
| Enrichment Score (ES) | -0.42831236 |
| Normalized Enrichment Score (NES) | -1.6611575 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.033961397 |
| FWER p-Value | 0.412 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AMD1 | 1448484_at | 56 | 7.018 | 0.0201 | No | ||
| 2 | TIMM10 | 1417499_at | 398 | 4.354 | 0.0186 | No | ||
| 3 | ERH | 1430536_a_at | 677 | 3.609 | 0.0175 | No | ||
| 4 | ACSL5 | 1428082_at | 712 | 3.538 | 0.0274 | No | ||
| 5 | HMGA1 | 1416184_s_at | 729 | 3.499 | 0.0379 | No | ||
| 6 | HIRIP3 | 1434936_at | 920 | 3.157 | 0.0394 | No | ||
| 7 | SIRT3 | 1417892_a_at | 986 | 3.070 | 0.0464 | No | ||
| 8 | SEMA4C | 1433920_at | 1091 | 2.954 | 0.0512 | No | ||
| 9 | SLC39A11 | 1424905_a_at | 1092 | 2.952 | 0.0607 | No | ||
| 10 | CCL2 | 1420380_at | 1140 | 2.887 | 0.0679 | No | ||
| 11 | CANX | 1415692_s_at 1422845_at 1428935_at | 1180 | 2.840 | 0.0753 | No | ||
| 12 | POLL | 1449352_at | 1243 | 2.771 | 0.0814 | No | ||
| 13 | PPIL1 | 1428892_at | 1665 | 2.383 | 0.0698 | No | ||
| 14 | TAF5 | 1435474_at | 1781 | 2.291 | 0.0719 | No | ||
| 15 | DPPA4 | 1429597_at | 1848 | 2.248 | 0.0762 | No | ||
| 16 | FXC1 | 1417916_a_at 1436416_x_at 1448887_x_at 1453461_at | 1902 | 2.217 | 0.0809 | No | ||
| 17 | BTAF1 | 1435248_a_at 1435249_at 1435953_at | 2260 | 2.020 | 0.0710 | No | ||
| 18 | LTBR | 1416435_at | 2401 | 1.951 | 0.0709 | No | ||
| 19 | LCP1 | 1415983_at 1448160_at | 2847 | 1.771 | 0.0562 | No | ||
| 20 | PLCG1 | 1435149_at 1450360_at | 2888 | 1.757 | 0.0601 | No | ||
| 21 | DIABLO | 1417683_at 1425768_at | 3046 | 1.700 | 0.0584 | No | ||
| 22 | PAFAH1B2 | 1422791_at 1422792_at 1422793_at 1450755_at | 3177 | 1.657 | 0.0578 | No | ||
| 23 | CLMN | 1425321_a_at 1436539_at 1439117_at 1451656_at 1457985_at | 3179 | 1.657 | 0.0631 | No | ||
| 24 | CPNE8 | 1430520_at 1430521_s_at 1431146_a_at | 3195 | 1.654 | 0.0677 | No | ||
| 25 | OGG1 | 1430078_a_at 1448815_at | 3262 | 1.635 | 0.0700 | No | ||
| 26 | CSAD | 1427981_a_at | 3350 | 1.607 | 0.0712 | No | ||
| 27 | STARD13 | 1452604_at | 3400 | 1.590 | 0.0741 | No | ||
| 28 | ADAMTS4 | 1452595_at | 3426 | 1.585 | 0.0781 | No | ||
| 29 | SOX14 | 1427609_at | 3469 | 1.569 | 0.0812 | No | ||
| 30 | CASP8 | 1424552_at | 3709 | 1.500 | 0.0751 | No | ||
| 31 | SART3 | 1417547_at 1417548_at | 3716 | 1.499 | 0.0797 | No | ||
| 32 | ELK4 | 1422233_at 1427162_a_at 1447571_at 1450549_s_at | 3803 | 1.475 | 0.0805 | No | ||
| 33 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 3962 | 1.430 | 0.0779 | No | ||
| 34 | STX11 | 1423576_a_at 1451032_at 1453228_at 1457780_at | 4087 | 1.396 | 0.0767 | No | ||
| 35 | RIN1 | 1424507_at 1437460_x_at | 4275 | 1.358 | 0.0725 | No | ||
| 36 | FBXO22 | 1426592_a_at 1426593_a_at 1452121_at | 4338 | 1.342 | 0.0740 | No | ||
| 37 | GGTL3 | 1451094_at | 4728 | 1.251 | 0.0602 | No | ||
| 38 | RNF12 | 1417250_at 1418730_at 1418731_at 1439403_x_at 1440850_at 1443167_at | 4911 | 1.217 | 0.0558 | No | ||
| 39 | RHOV | 1424976_at | 4952 | 1.209 | 0.0578 | No | ||
| 40 | CAST | 1426098_a_at 1435972_at 1451413_at | 4983 | 1.201 | 0.0603 | No | ||
| 41 | C1QTNF6 | 1431856_a_at | 5434 | 1.120 | 0.0433 | No | ||
| 42 | USP3 | 1425022_at 1425023_at 1435531_at 1441056_at 1446886_at | 5447 | 1.117 | 0.0464 | No | ||
| 43 | DCLRE1C | 1457679_at 1458361_at | 5663 | 1.079 | 0.0400 | No | ||
| 44 | FIBP | 1422516_a_at 1430171_at 1438923_at 1438924_x_at | 5794 | 1.057 | 0.0374 | No | ||
| 45 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 6317 | 0.979 | 0.0166 | No | ||
| 46 | ARHGAP4 | 1419296_at 1443360_x_at 1458190_at | 6334 | 0.976 | 0.0191 | No | ||
| 47 | UGCGL2 | 1429836_at 1431602_a_at | 6492 | 0.953 | 0.0149 | No | ||
| 48 | LIN28 | 1437752_at | 6538 | 0.947 | 0.0159 | No | ||
| 49 | SHC1 | 1422853_at 1422854_at | 6567 | 0.943 | 0.0177 | No | ||
| 50 | NR4A2 | 1447863_s_at 1450749_a_at 1450750_a_at 1455034_at | 6999 | 0.875 | 0.0007 | No | ||
| 51 | EXTL2 | 1422538_at 1422539_at | 7163 | 0.849 | -0.0040 | No | ||
| 52 | MARK1 | 1419834_x_at 1425509_at 1425510_at 1425511_at 1449630_s_at | 7270 | 0.830 | -0.0062 | No | ||
| 53 | CD79A | 1418830_at | 7368 | 0.816 | -0.0080 | No | ||
| 54 | UBE2N | 1422559_at 1435384_at | 7699 | 0.767 | -0.0207 | No | ||
| 55 | SPIB | 1460407_at | 7760 | 0.760 | -0.0210 | No | ||
| 56 | HOXC4 | 1422870_at | 7807 | 0.753 | -0.0206 | No | ||
| 57 | PNMA1 | 1429224_at | 7907 | 0.737 | -0.0228 | No | ||
| 58 | FXYD5 | 1418296_at | 9121 | 0.557 | -0.0767 | No | ||
| 59 | DES | 1426731_at | 9319 | 0.529 | -0.0840 | No | ||
| 60 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 9382 | 0.522 | -0.0852 | No | ||
| 61 | TENC1 | 1452264_at | 9987 | 0.437 | -0.1115 | No | ||
| 62 | CGGBP1 | 1454641_at 1455658_at | 10117 | 0.421 | -0.1161 | No | ||
| 63 | NXT2 | 1456599_at | 10140 | 0.418 | -0.1157 | No | ||
| 64 | VAMP8 | 1420624_a_at | 10356 | 0.390 | -0.1243 | No | ||
| 65 | MGAT4A | 1444347_at | 10844 | 0.320 | -0.1457 | No | ||
| 66 | PSMD13 | 1422459_a_at 1447843_at | 10896 | 0.313 | -0.1470 | No | ||
| 67 | JPH4 | 1439031_at | 11489 | 0.228 | -0.1734 | No | ||
| 68 | TUSC3 | 1421662_a_at 1458568_at | 11520 | 0.223 | -0.1741 | No | ||
| 69 | SLC30A7 | 1450697_at | 11633 | 0.208 | -0.1786 | No | ||
| 70 | RNPS1 | 1437027_x_at 1437359_at 1437851_x_at 1438267_x_at 1448324_at | 11722 | 0.194 | -0.1820 | No | ||
| 71 | CAPZA1 | 1439455_x_at 1440380_at | 12277 | 0.108 | -0.2071 | No | ||
| 72 | DDX50 | 1417875_at 1439082_at | 12307 | 0.104 | -0.2081 | No | ||
| 73 | GRB7 | 1448227_at | 12679 | 0.051 | -0.2249 | No | ||
| 74 | CTSS | 1448591_at | 12868 | 0.023 | -0.2335 | No | ||
| 75 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 13113 | -0.022 | -0.2446 | No | ||
| 76 | ARHGAP15 | 1429881_at 1435959_at 1441650_at | 13285 | -0.052 | -0.2523 | No | ||
| 77 | RIN3 | 1434684_at 1442530_at | 13345 | -0.062 | -0.2548 | No | ||
| 78 | CORO1C | 1417752_at 1419911_at 1437721_at 1442972_at 1449660_s_at 1459169_at | 13396 | -0.070 | -0.2569 | No | ||
| 79 | PRKCBP1 | 1426614_at 1429415_at 1445827_at 1456143_at 1458617_at | 14038 | -0.185 | -0.2857 | No | ||
| 80 | HYAL2 | 1448679_at | 14149 | -0.204 | -0.2901 | No | ||
| 81 | CHES1 | 1434002_at 1436925_at 1438255_at 1440607_at 1444555_at 1447325_at 1453094_at | 15039 | -0.376 | -0.3297 | No | ||
| 82 | IL23A | 1419529_at | 15040 | -0.376 | -0.3285 | No | ||
| 83 | TEAD3 | 1420593_a_at | 15325 | -0.435 | -0.3401 | No | ||
| 84 | ING4 | 1448054_at 1460728_s_at | 15551 | -0.496 | -0.3489 | No | ||
| 85 | SCAMP2 | 1416611_at 1448404_at | 15568 | -0.502 | -0.3480 | No | ||
| 86 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 15690 | -0.533 | -0.3518 | No | ||
| 87 | MUSK | 1450511_at | 15782 | -0.551 | -0.3542 | No | ||
| 88 | ST7L | 1427916_at 1456363_at | 15869 | -0.569 | -0.3563 | No | ||
| 89 | POU3F4 | 1422164_at 1422165_at | 15994 | -0.600 | -0.3601 | No | ||
| 90 | NDUFS2 | 1451096_at | 16183 | -0.653 | -0.3666 | No | ||
| 91 | SLC25A5 | 1423772_x_at 1434801_x_at 1436874_x_at 1445944_at | 16344 | -0.700 | -0.3717 | No | ||
| 92 | U2AF2 | 1417260_at | 16618 | -0.781 | -0.3817 | No | ||
| 93 | SNRPB | 1419260_a_at 1437193_s_at | 16788 | -0.837 | -0.3867 | No | ||
| 94 | BIN3 | 1417691_at | 16960 | -0.887 | -0.3917 | No | ||
| 95 | PRKACA | 1447720_x_at 1450519_a_at | 16985 | -0.895 | -0.3899 | No | ||
| 96 | LYPLA2 | 1417433_at | 17358 | -1.018 | -0.4037 | No | ||
| 97 | NEDD8 | 1423715_a_at 1440996_at 1445811_at | 17362 | -1.019 | -0.4005 | No | ||
| 98 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 17364 | -1.019 | -0.3973 | No | ||
| 99 | MRPL52 | 1415761_at 1415762_x_at 1455366_at 1460701_a_at | 17377 | -1.023 | -0.3945 | No | ||
| 100 | ERCC6 | 1441595_at 1442604_at | 17408 | -1.033 | -0.3926 | No | ||
| 101 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 17409 | -1.033 | -0.3892 | No | ||
| 102 | DMTF1 | 1422636_at 1440349_at 1456697_x_at | 17465 | -1.050 | -0.3884 | No | ||
| 103 | NR1D1 | 1421802_at 1426464_at | 17482 | -1.055 | -0.3857 | No | ||
| 104 | TBN | 1416450_at 1416451_s_at | 17713 | -1.161 | -0.3925 | No | ||
| 105 | RNF6 | 1427898_at 1427899_at | 17734 | -1.169 | -0.3896 | No | ||
| 106 | CD2BP2 | 1417223_at 1417224_a_at 1448624_at | 17812 | -1.198 | -0.3893 | No | ||
| 107 | HM13 | 1417287_at 1428855_at 1428856_at 1438456_at 1438865_at 1455631_at | 18009 | -1.278 | -0.3941 | No | ||
| 108 | PRKACB | 1420610_at 1420611_at 1457197_at | 18425 | -1.475 | -0.4084 | No | ||
| 109 | DPP3 | 1431594_at 1448215_a_at | 18459 | -1.493 | -0.4051 | No | ||
| 110 | TGIF2 | 1431115_at 1434400_at | 18700 | -1.598 | -0.4110 | No | ||
| 111 | SEC24C | 1430250_at 1452147_at | 19079 | -1.846 | -0.4223 | Yes | ||
| 112 | FCHO1 | 1428575_at 1436077_a_at 1436078_at | 19170 | -1.916 | -0.4203 | Yes | ||
| 113 | STAT4 | 1448713_at | 19172 | -1.918 | -0.4141 | Yes | ||
| 114 | SNX1 | 1416260_a_at | 19339 | -2.037 | -0.4151 | Yes | ||
| 115 | RALB | 1417744_a_at 1435517_x_at | 19408 | -2.076 | -0.4116 | Yes | ||
| 116 | DDIT3 | 1417516_at 1443897_at | 19630 | -2.223 | -0.4145 | Yes | ||
| 117 | CHD2 | 1444246_at 1445843_at | 19671 | -2.253 | -0.4091 | Yes | ||
| 118 | HTATIP | 1433980_at 1433981_s_at | 19695 | -2.276 | -0.4027 | Yes | ||
| 119 | ARRB2 | 1426239_s_at 1451987_at | 19731 | -2.311 | -0.3969 | Yes | ||
| 120 | HERC4 | 1427032_at 1431716_at 1456677_at | 19746 | -2.321 | -0.3900 | Yes | ||
| 121 | AGL | 1429083_at 1431032_at 1431033_x_at | 19749 | -2.326 | -0.3826 | Yes | ||
| 122 | ESRRA | 1442864_at 1460652_at | 19955 | -2.536 | -0.3838 | Yes | ||
| 123 | ARHGAP1 | 1424307_at 1451309_at | 19996 | -2.579 | -0.3773 | Yes | ||
| 124 | FURIN | 1418518_at | 20066 | -2.653 | -0.3719 | Yes | ||
| 125 | CRSP7 | 1452282_at 1458377_at | 20069 | -2.656 | -0.3634 | Yes | ||
| 126 | PPP4C | 1445350_at 1460288_a_at | 20181 | -2.780 | -0.3595 | Yes | ||
| 127 | CD3E | 1422105_at 1445748_at | 20189 | -2.789 | -0.3508 | Yes | ||
| 128 | E2F5 | 1417444_at 1447625_at 1460207_s_at | 20228 | -2.832 | -0.3433 | Yes | ||
| 129 | PTPN6 | 1456694_x_at 1460188_at | 20280 | -2.884 | -0.3364 | Yes | ||
| 130 | AGPAT1 | 1421024_at 1421025_at | 20286 | -2.887 | -0.3272 | Yes | ||
| 131 | ZDHHC5 | 1426310_at 1426311_s_at | 20332 | -2.956 | -0.3197 | Yes | ||
| 132 | TRAPPC1 | 1436019_a_at | 20389 | -3.015 | -0.3126 | Yes | ||
| 133 | DAPP1 | 1421936_at 1421937_at 1447337_at | 20602 | -3.311 | -0.3116 | Yes | ||
| 134 | ARHGEF7 | 1424482_at 1449066_a_at | 20618 | -3.335 | -0.3015 | Yes | ||
| 135 | PIK3R4 | 1460352_s_at | 20620 | -3.336 | -0.2907 | Yes | ||
| 136 | CENTG1 | 1419789_at | 20801 | -3.605 | -0.2873 | Yes | ||
| 137 | MAP3K11 | 1450669_at | 20827 | -3.653 | -0.2767 | Yes | ||
| 138 | GMPR2 | 1416356_at 1429180_at | 20864 | -3.710 | -0.2663 | Yes | ||
| 139 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 20871 | -3.716 | -0.2546 | Yes | ||
| 140 | CENTD2 | 1428064_at | 20965 | -3.855 | -0.2464 | Yes | ||
| 141 | MARK3 | 1418316_a_at | 20967 | -3.857 | -0.2339 | Yes | ||
| 142 | TMEM24 | 1428095_a_at 1457585_at | 20968 | -3.859 | -0.2215 | Yes | ||
| 143 | HCLS1 | 1418842_at | 21024 | -3.955 | -0.2112 | Yes | ||
| 144 | DGKA | 1418578_at | 21145 | -4.248 | -0.2030 | Yes | ||
| 145 | TRIM41 | 1424275_s_at 1440268_at | 21245 | -4.511 | -0.1929 | Yes | ||
| 146 | ARFIP2 | 1424240_at 1435498_at | 21356 | -4.802 | -0.1824 | Yes | ||
| 147 | ZFP95 | 1424892_at 1430562_at | 21388 | -4.889 | -0.1680 | Yes | ||
| 148 | CAP1 | 1417461_at 1417462_at | 21432 | -5.083 | -0.1536 | Yes | ||
| 149 | SLC9A3R1 | 1438115_a_at 1438116_x_at 1450982_at | 21454 | -5.193 | -0.1377 | Yes | ||
| 150 | SIPA1 | 1416206_at | 21594 | -5.880 | -0.1251 | Yes | ||
| 151 | JUNB | 1415899_at | 21600 | -5.913 | -0.1062 | Yes | ||
| 152 | GFI1 | 1417679_at | 21682 | -6.457 | -0.0890 | Yes | ||
| 153 | DNAJC14 | 1426553_at 1437546_at 1437547_s_at | 21705 | -6.772 | -0.0681 | Yes | ||
| 154 | CTSW | 1422632_at | 21733 | -7.037 | -0.0466 | Yes | ||
| 155 | RGS14 | 1419221_a_at | 21805 | -8.252 | -0.0231 | Yes | ||
| 156 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21845 | -8.970 | 0.0041 | Yes |

