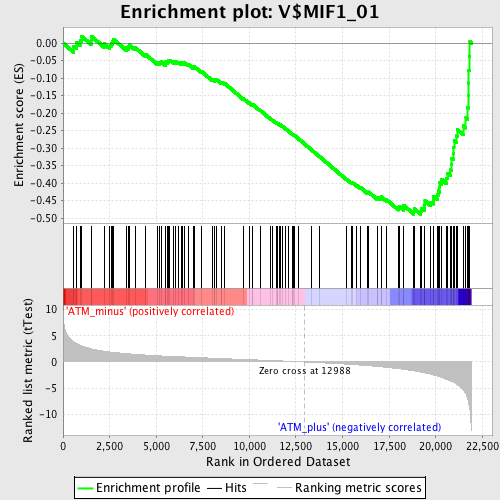
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$MIF1_01 |
| Enrichment Score (ES) | -0.48954412 |
| Normalized Enrichment Score (NES) | -1.8097816 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011196029 |
| FWER p-Value | 0.068 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHCHD4 | 1417233_at | 576 | 3.799 | -0.0082 | No | ||
| 2 | HAT1 | 1428061_at | 716 | 3.532 | 0.0023 | No | ||
| 3 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 909 | 3.180 | 0.0088 | No | ||
| 4 | GRID2 | 1421435_at 1421436_at 1435487_at 1437824_at 1443101_at 1446879_at 1458756_at 1459245_s_at | 982 | 3.076 | 0.0202 | No | ||
| 5 | FGR | 1419526_at 1442804_at | 1502 | 2.514 | 0.0085 | No | ||
| 6 | CARS | 1427330_at 1452394_at | 1511 | 2.509 | 0.0201 | No | ||
| 7 | TPCN1 | 1434930_at 1451771_at 1451772_at | 2199 | 2.047 | -0.0016 | No | ||
| 8 | CRLF1 | 1418476_at | 2516 | 1.898 | -0.0070 | No | ||
| 9 | PLA2G12A | 1452026_a_at | 2574 | 1.869 | -0.0006 | No | ||
| 10 | NEK2 | 1417299_at 1437579_at 1437580_s_at 1443999_at | 2635 | 1.848 | 0.0055 | No | ||
| 11 | FIBCD1 | 1435482_at | 2691 | 1.827 | 0.0117 | No | ||
| 12 | PTCH2 | 1422655_at | 3386 | 1.593 | -0.0125 | No | ||
| 13 | MDH1B | 1429842_at | 3491 | 1.564 | -0.0097 | No | ||
| 14 | GALNT14 | 1453630_at | 3550 | 1.546 | -0.0050 | No | ||
| 15 | EMX2 | 1456258_at | 3860 | 1.455 | -0.0122 | No | ||
| 16 | TLX3 | 1450806_at | 4409 | 1.324 | -0.0310 | No | ||
| 17 | TTC16 | 1455414_at | 5064 | 1.187 | -0.0552 | No | ||
| 18 | SOX3 | 1435192_at 1450485_at | 5175 | 1.168 | -0.0547 | No | ||
| 19 | GRIK5 | 1418784_at | 5262 | 1.150 | -0.0531 | No | ||
| 20 | TEKT2 | 1418484_at | 5496 | 1.108 | -0.0585 | No | ||
| 21 | HMGCS1 | 1433443_a_at 1433444_at 1433445_x_at 1433446_at 1441536_at | 5507 | 1.106 | -0.0537 | No | ||
| 22 | MYCBP | 1421165_at 1427938_at 1427939_s_at 1427940_s_at 1452608_at | 5621 | 1.086 | -0.0536 | No | ||
| 23 | LRP2 | 1427133_s_at 1427621_at 1445901_at 1452320_at | 5668 | 1.079 | -0.0506 | No | ||
| 24 | PLCB3 | 1448661_at | 5728 | 1.067 | -0.0482 | No | ||
| 25 | LSM7 | 1417313_at | 5943 | 1.036 | -0.0530 | No | ||
| 26 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 6056 | 1.017 | -0.0533 | No | ||
| 27 | WARS2 | 1429279_at 1435273_at | 6191 | 0.999 | -0.0546 | No | ||
| 28 | TTC8 | 1424410_at | 6358 | 0.973 | -0.0576 | No | ||
| 29 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 6384 | 0.968 | -0.0541 | No | ||
| 30 | BMP6 | 1450759_at 1459947_at | 6508 | 0.952 | -0.0552 | No | ||
| 31 | TBC1D16 | 1435763_at 1442149_at | 6722 | 0.916 | -0.0605 | No | ||
| 32 | RNF41 | 1423842_a_at 1432003_a_at 1453633_a_at 1455763_at | 7003 | 0.874 | -0.0692 | No | ||
| 33 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 7052 | 0.866 | -0.0672 | No | ||
| 34 | GLRA3 | 1450239_at | 7429 | 0.806 | -0.0806 | No | ||
| 35 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 8003 | 0.724 | -0.1034 | No | ||
| 36 | GRM3 | 1430136_at | 8119 | 0.707 | -0.1053 | No | ||
| 37 | ARL4A | 1425411_at 1431429_a_at 1435092_at | 8143 | 0.702 | -0.1030 | No | ||
| 38 | MAGED1 | 1450062_a_at | 8230 | 0.688 | -0.1036 | No | ||
| 39 | CSMD3 | 1438272_at 1438572_at 1454245_at | 8501 | 0.648 | -0.1129 | No | ||
| 40 | TGFB3 | 1417455_at | 8527 | 0.644 | -0.1109 | No | ||
| 41 | WWP1 | 1427097_at 1427098_at 1446364_at 1446853_at 1452299_at | 8692 | 0.619 | -0.1155 | No | ||
| 42 | SNF1LK | 1419766_at 1459601_at | 9692 | 0.478 | -0.1590 | No | ||
| 43 | SMAD5 | 1421047_at 1433641_at 1451873_a_at | 9993 | 0.436 | -0.1706 | No | ||
| 44 | PTDSS2 | 1426728_x_at 1429492_x_at 1434586_a_at 1434587_x_at 1439984_at 1443738_at 1450354_a_at 1453164_a_at 1458387_at 1460010_a_at | 10144 | 0.418 | -0.1755 | No | ||
| 45 | NELF | 1436959_x_at 1439398_x_at 1442706_at 1449474_a_at | 10180 | 0.414 | -0.1751 | No | ||
| 46 | DNAH9 | 1442412_at 1457401_at | 10578 | 0.355 | -0.1916 | No | ||
| 47 | GPRC5D | 1420538_at | 11159 | 0.277 | -0.2169 | No | ||
| 48 | SPA17 | 1417635_at | 11270 | 0.260 | -0.2207 | No | ||
| 49 | EFNB3 | 1423085_at | 11452 | 0.232 | -0.2279 | No | ||
| 50 | TUSC3 | 1421662_a_at 1458568_at | 11520 | 0.223 | -0.2299 | No | ||
| 51 | MRPL10 | 1418112_at | 11538 | 0.221 | -0.2296 | No | ||
| 52 | PRKCSH | 1416339_a_at | 11640 | 0.206 | -0.2332 | No | ||
| 53 | SDCBP2 | 1424090_at | 11655 | 0.204 | -0.2329 | No | ||
| 54 | NAT5 | 1418244_at 1422961_at | 11775 | 0.186 | -0.2374 | No | ||
| 55 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 11962 | 0.157 | -0.2452 | No | ||
| 56 | STMN4 | 1418105_at | 12094 | 0.137 | -0.2506 | No | ||
| 57 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 12302 | 0.105 | -0.2595 | No | ||
| 58 | MDH1 | 1434319_at 1436834_x_at 1438338_at 1448172_at 1454925_x_at | 12338 | 0.100 | -0.2607 | No | ||
| 59 | PACRG | 1428751_at | 12372 | 0.095 | -0.2617 | No | ||
| 60 | TBPL1 | 1431415_a_at 1434907_at | 12414 | 0.089 | -0.2632 | No | ||
| 61 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 12645 | 0.057 | -0.2734 | No | ||
| 62 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 13357 | -0.063 | -0.3057 | No | ||
| 63 | FILIP1 | 1436650_at 1444075_at | 13769 | -0.137 | -0.3239 | No | ||
| 64 | CPEB2 | 1434272_at 1443017_at 1458518_at | 15227 | -0.412 | -0.3887 | No | ||
| 65 | VEGF | 1420909_at 1451959_a_at | 15473 | -0.473 | -0.3977 | No | ||
| 66 | BBS2 | 1424478_at | 15527 | -0.489 | -0.3977 | No | ||
| 67 | ARID1A | 1447104_at | 15769 | -0.549 | -0.4062 | No | ||
| 68 | PARK2 | 1420755_a_at 1426135_a_at 1449975_a_at | 15958 | -0.590 | -0.4119 | No | ||
| 69 | THNSL1 | 1427936_at 1452605_at | 16360 | -0.704 | -0.4270 | No | ||
| 70 | ITGB8 | 1436223_at | 16396 | -0.713 | -0.4251 | No | ||
| 71 | ADCK4 | 1448490_at | 16867 | -0.859 | -0.4426 | No | ||
| 72 | GTF3C4 | 1434320_at 1438692_at | 16900 | -0.869 | -0.4399 | No | ||
| 73 | DPAGT1 | 1448549_a_at | 17076 | -0.928 | -0.4434 | No | ||
| 74 | NRGN | 1423231_at | 17080 | -0.930 | -0.4391 | No | ||
| 75 | NEDD8 | 1423715_a_at 1440996_at 1445811_at | 17362 | -1.019 | -0.4471 | No | ||
| 76 | HM13 | 1417287_at 1428855_at 1428856_at 1438456_at 1438865_at 1455631_at | 18009 | -1.278 | -0.4706 | No | ||
| 77 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 18076 | -1.306 | -0.4674 | No | ||
| 78 | CBX6 | 1424407_s_at 1429290_at | 18293 | -1.413 | -0.4705 | No | ||
| 79 | PHF15 | 1430081_at 1455345_at | 18295 | -1.413 | -0.4638 | No | ||
| 80 | CBLB | 1437304_at 1455082_at 1458469_at | 18835 | -1.687 | -0.4804 | Yes | ||
| 81 | NCDN | 1416852_a_at 1416853_at | 18844 | -1.689 | -0.4727 | Yes | ||
| 82 | STK36 | 1434733_at 1453584_at | 19213 | -1.956 | -0.4802 | Yes | ||
| 83 | DNAJC1 | 1420500_at 1420501_at 1440551_at 1447649_x_at 1453372_at 1459791_at | 19229 | -1.966 | -0.4715 | Yes | ||
| 84 | RASGEF1A | 1429316_at 1435218_at | 19402 | -2.070 | -0.4694 | Yes | ||
| 85 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 19416 | -2.083 | -0.4600 | Yes | ||
| 86 | POLD4 | 1427885_at | 19431 | -2.091 | -0.4507 | Yes | ||
| 87 | RFX2 | 1417332_at | 19751 | -2.329 | -0.4541 | Yes | ||
| 88 | KIF3B | 1420987_at 1450073_at 1450074_at | 19873 | -2.449 | -0.4480 | Yes | ||
| 89 | FGF13 | 1418497_at 1418498_at | 19892 | -2.472 | -0.4369 | Yes | ||
| 90 | SALL2 | 1416638_at | 20090 | -2.690 | -0.4331 | Yes | ||
| 91 | KIF17 | 1419826_at 1419827_s_at 1422762_at | 20137 | -2.734 | -0.4221 | Yes | ||
| 92 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 20218 | -2.820 | -0.4123 | Yes | ||
| 93 | E2F5 | 1417444_at 1447625_at 1460207_s_at | 20228 | -2.832 | -0.3991 | Yes | ||
| 94 | DCTN1 | 1422521_at 1435414_s_at | 20326 | -2.947 | -0.3894 | Yes | ||
| 95 | AP1M1 | 1416307_at 1459832_s_at | 20582 | -3.295 | -0.3853 | Yes | ||
| 96 | NEDD9 | 1422818_at 1437132_x_at 1447885_x_at 1450767_at | 20631 | -3.353 | -0.3715 | Yes | ||
| 97 | BDNF | 1422168_a_at 1422169_a_at | 20807 | -3.621 | -0.3622 | Yes | ||
| 98 | GMPR2 | 1416356_at 1429180_at | 20864 | -3.710 | -0.3470 | Yes | ||
| 99 | SREBF1 | 1426690_a_at | 20873 | -3.719 | -0.3295 | Yes | ||
| 100 | CCDC6 | 1428311_at 1459159_a_at | 20950 | -3.824 | -0.3147 | Yes | ||
| 101 | TMEM24 | 1428095_a_at 1457585_at | 20968 | -3.859 | -0.2970 | Yes | ||
| 102 | PSCD2 | 1450103_a_at | 20991 | -3.906 | -0.2793 | Yes | ||
| 103 | TRIM39 | 1418851_at | 21110 | -4.153 | -0.2648 | Yes | ||
| 104 | NCOA5 | 1424055_at 1456997_at | 21169 | -4.305 | -0.2469 | Yes | ||
| 105 | SOCS1 | 1440047_at 1450446_a_at | 21486 | -5.335 | -0.2358 | Yes | ||
| 106 | MADD | 1425735_at 1455502_at | 21632 | -6.088 | -0.2133 | Yes | ||
| 107 | RNF25 | 1442108_at 1448674_at | 21732 | -7.036 | -0.1841 | Yes | ||
| 108 | CBX4 | 1419583_at 1440479_at | 21761 | -7.444 | -0.1497 | Yes | ||
| 109 | DDX31 | 1434182_at 1438962_s_at | 21766 | -7.554 | -0.1138 | Yes | ||
| 110 | SPRY2 | 1421656_at 1436584_at | 21772 | -7.739 | -0.0769 | Yes | ||
| 111 | DHRS3 | 1448390_a_at | 21826 | -8.596 | -0.0382 | Yes | ||
| 112 | CCRK | 1422494_s_at | 21846 | -8.997 | 0.0040 | Yes |

