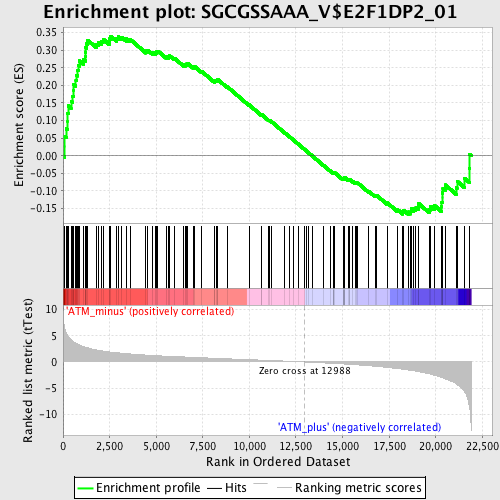
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | SGCGSSAAA_V$E2F1DP2_01 |
| Enrichment Score (ES) | 0.3399115 |
| Normalized Enrichment Score (NES) | 1.4140368 |
| Nominal p-value | 0.009063444 |
| FDR q-value | 0.12795709 |
| FWER p-Value | 0.937 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MCM4 | 1416214_at 1436708_x_at | 94 | 6.274 | 0.0251 | Yes | ||
| 2 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 95 | 6.259 | 0.0545 | Yes | ||
| 3 | MYC | 1424942_a_at | 165 | 5.495 | 0.0771 | Yes | ||
| 4 | PRPS1 | 1416052_at | 244 | 5.034 | 0.0972 | Yes | ||
| 5 | SNRPD1 | 1416336_s_at | 250 | 5.006 | 0.1204 | Yes | ||
| 6 | ATF5 | 1425927_a_at | 280 | 4.852 | 0.1418 | Yes | ||
| 7 | TOPBP1 | 1452241_at 1458480_at | 429 | 4.243 | 0.1550 | Yes | ||
| 8 | DNAJC9 | 1426473_at | 517 | 3.954 | 0.1695 | Yes | ||
| 9 | PHF5A | 1424170_at | 563 | 3.833 | 0.1855 | Yes | ||
| 10 | RANBP1 | 1422547_at | 570 | 3.814 | 0.2031 | Yes | ||
| 11 | GMNN | 1417506_at | 688 | 3.583 | 0.2145 | Yes | ||
| 12 | HMGA1 | 1416184_s_at | 729 | 3.499 | 0.2291 | Yes | ||
| 13 | FKBP5 | 1416125_at 1447736_at 1448231_at 1458089_at | 789 | 3.389 | 0.2423 | Yes | ||
| 14 | UNG | 1425753_a_at | 811 | 3.351 | 0.2571 | Yes | ||
| 15 | RQCD1 | 1450887_at | 869 | 3.249 | 0.2697 | Yes | ||
| 16 | POLE4 | 1423371_at 1423372_at 1432286_at 1444739_at | 1100 | 2.946 | 0.2730 | Yes | ||
| 17 | CDC25A | 1417131_at 1417132_at 1445995_at | 1192 | 2.824 | 0.2820 | Yes | ||
| 18 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1203 | 2.812 | 0.2948 | Yes | ||
| 19 | SUV39H1 | 1427382_a_at 1432227_at 1432236_a_at | 1216 | 2.803 | 0.3074 | Yes | ||
| 20 | CDCA7 | 1428069_at 1445681_at | 1268 | 2.749 | 0.3179 | Yes | ||
| 21 | PCNA | 1417947_at | 1322 | 2.702 | 0.3282 | Yes | ||
| 22 | MRPL40 | 1448849_at | 1796 | 2.281 | 0.3172 | Yes | ||
| 23 | SLC9A5 | 1420070_a_at 1455760_at | 1907 | 2.216 | 0.3226 | Yes | ||
| 24 | E2F3 | 1427462_at 1434564_at | 2059 | 2.126 | 0.3256 | Yes | ||
| 25 | AK2 | 1448450_at 1448451_at | 2147 | 2.072 | 0.3314 | Yes | ||
| 26 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 2498 | 1.902 | 0.3242 | Yes | ||
| 27 | PTMA | 1423455_at | 2509 | 1.898 | 0.3327 | Yes | ||
| 28 | HNRPR | 1427129_a_at 1438807_at 1452030_a_at | 2545 | 1.882 | 0.3399 | Yes | ||
| 29 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 2868 | 1.765 | 0.3334 | No | ||
| 30 | POLE2 | 1427094_at 1431440_at | 2947 | 1.735 | 0.3380 | No | ||
| 31 | NRK | 1436399_s_at 1450078_at 1450079_at | 3138 | 1.668 | 0.3371 | No | ||
| 32 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 3406 | 1.589 | 0.3323 | No | ||
| 33 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 3592 | 1.530 | 0.3310 | No | ||
| 34 | KBTBD7 | 1428192_at 1439865_at 1452700_s_at | 4450 | 1.311 | 0.2979 | No | ||
| 35 | MSH5 | 1430771_a_at 1449537_at | 4541 | 1.291 | 0.2998 | No | ||
| 36 | FBXO5 | 1429499_at | 4803 | 1.237 | 0.2937 | No | ||
| 37 | PKMYT1 | 1418481_at 1438601_at | 4959 | 1.207 | 0.2922 | No | ||
| 38 | STAG2 | 1421849_at 1450396_at | 5019 | 1.195 | 0.2951 | No | ||
| 39 | PRPF4B | 1425497_a_at 1425498_at 1436427_at 1451732_at 1451909_a_at 1455696_a_at | 5090 | 1.184 | 0.2975 | No | ||
| 40 | POLD1 | 1448187_at 1456055_x_at | 5538 | 1.101 | 0.2822 | No | ||
| 41 | MCM3 | 1420029_at 1426653_at | 5651 | 1.081 | 0.2821 | No | ||
| 42 | ARHGAP6 | 1417704_a_at 1451867_x_at 1456333_a_at | 5706 | 1.071 | 0.2847 | No | ||
| 43 | GPRC5B | 1424613_at 1451411_at | 5972 | 1.030 | 0.2773 | No | ||
| 44 | SFRS7 | 1424033_at 1424883_s_at 1436871_at | 6480 | 0.955 | 0.2586 | No | ||
| 45 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 6566 | 0.943 | 0.2591 | No | ||
| 46 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 6626 | 0.932 | 0.2608 | No | ||
| 47 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 6672 | 0.924 | 0.2631 | No | ||
| 48 | CLSPN | 1441984_at 1456280_at | 7017 | 0.872 | 0.2514 | No | ||
| 49 | PAQR4 | 1423101_at | 7051 | 0.867 | 0.2539 | No | ||
| 50 | GLRA3 | 1450239_at | 7429 | 0.806 | 0.2404 | No | ||
| 51 | GABRB3 | 1421189_at 1421190_at 1435021_at | 8137 | 0.703 | 0.2113 | No | ||
| 52 | PPM1D | 1434142_at 1449092_at 1459069_at | 8145 | 0.702 | 0.2143 | No | ||
| 53 | PCSK1 | 1421396_at | 8238 | 0.687 | 0.2133 | No | ||
| 54 | BRMS1L | 1452850_s_at 1460443_at | 8273 | 0.683 | 0.2150 | No | ||
| 55 | H2AFV | 1428029_a_at 1436596_at | 8307 | 0.677 | 0.2166 | No | ||
| 56 | RET | 1421359_at | 8822 | 0.600 | 0.1959 | No | ||
| 57 | PIM1 | 1423006_at 1435458_at | 9985 | 0.437 | 0.1447 | No | ||
| 58 | FANCC | 1442265_at 1447632_at 1450861_at | 10628 | 0.347 | 0.1169 | No | ||
| 59 | DNMT1 | 1422946_a_at 1435122_x_at 1445637_at 1447877_x_at | 10638 | 0.346 | 0.1181 | No | ||
| 60 | NR6A1 | 1421515_at 1421516_at 1428825_at 1428826_at 1445872_at | 11053 | 0.293 | 0.1005 | No | ||
| 61 | ASXL2 | 1439063_at 1444754_at 1453295_at 1458366_at 1460597_at | 11094 | 0.287 | 0.1000 | No | ||
| 62 | GATA1 | 1449232_at | 11198 | 0.270 | 0.0965 | No | ||
| 63 | AP4M1 | 1418846_at | 11901 | 0.166 | 0.0651 | No | ||
| 64 | KCNS2 | 1421342_at 1441211_at 1457325_at | 12165 | 0.126 | 0.0537 | No | ||
| 65 | ING3 | 1422805_a_at 1422806_x_at 1450760_a_at 1460082_at | 12384 | 0.094 | 0.0441 | No | ||
| 66 | PODN | 1435180_at | 12634 | 0.058 | 0.0330 | No | ||
| 67 | SYNGR4 | 1418875_at | 12953 | 0.006 | 0.0184 | No | ||
| 68 | PLAGL1 | 1426208_x_at 1438588_at 1450533_a_at | 13063 | -0.014 | 0.0135 | No | ||
| 69 | MSH2 | 1416988_at 1447888_x_at | 13177 | -0.033 | 0.0085 | No | ||
| 70 | ADAMTS2 | 1435990_at 1455720_at 1457058_at | 13373 | -0.065 | -0.0001 | No | ||
| 71 | ILF3 | 1422546_at 1436802_at 1460669_at | 13981 | -0.175 | -0.0271 | No | ||
| 72 | EED | 1448653_at | 14356 | -0.244 | -0.0431 | No | ||
| 73 | RPS20 | 1456373_x_at 1456436_x_at | 14512 | -0.273 | -0.0490 | No | ||
| 74 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 14535 | -0.276 | -0.0487 | No | ||
| 75 | HCN3 | 1451023_at | 14545 | -0.278 | -0.0478 | No | ||
| 76 | HIRA | 1436241_s_at 1450049_a_at 1450050_at 1459103_at | 14552 | -0.278 | -0.0468 | No | ||
| 77 | FHOD1 | 1435557_at | 15035 | -0.374 | -0.0671 | No | ||
| 78 | CDC6 | 1417019_a_at | 15043 | -0.376 | -0.0657 | No | ||
| 79 | FMO4 | 1425799_at 1444852_at | 15078 | -0.381 | -0.0654 | No | ||
| 80 | SFRS10 | 1419543_a_at 1427432_a_at 1441466_at 1446496_at | 15092 | -0.386 | -0.0642 | No | ||
| 81 | HOXC10 | 1439798_at | 15095 | -0.386 | -0.0625 | No | ||
| 82 | KCNA6 | 1421568_at 1441049_at 1456954_at | 15118 | -0.390 | -0.0617 | No | ||
| 83 | MCM2 | 1434079_s_at 1448777_at | 15310 | -0.431 | -0.0684 | No | ||
| 84 | TYRO3 | 1425248_a_at 1425249_a_at | 15357 | -0.445 | -0.0684 | No | ||
| 85 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 15380 | -0.452 | -0.0673 | No | ||
| 86 | PRKDC | 1451576_at | 15548 | -0.496 | -0.0726 | No | ||
| 87 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 15690 | -0.533 | -0.0766 | No | ||
| 88 | EFNA5 | 1421796_a_at 1436866_at 1440052_at 1446383_at 1451930_at | 15730 | -0.543 | -0.0758 | No | ||
| 89 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 15820 | -0.560 | -0.0773 | No | ||
| 90 | ATAD2 | 1436174_at 1443146_at 1443229_at | 16401 | -0.715 | -0.1005 | No | ||
| 91 | RAVER1 | 1427110_at 1427111_s_at 1444966_at | 16760 | -0.826 | -0.1131 | No | ||
| 92 | AP1S1 | 1416087_at | 16818 | -0.847 | -0.1117 | No | ||
| 93 | HNRPD | 1425142_a_at 1446083_at | 17398 | -1.029 | -0.1334 | No | ||
| 94 | HNRPUL1 | 1451984_at | 17966 | -1.263 | -0.1535 | No | ||
| 95 | NUP62 | 1415925_a_at 1415926_at 1437612_at 1438917_x_at 1447905_x_at 1448150_at | 18238 | -1.380 | -0.1594 | No | ||
| 96 | PHF15 | 1430081_at 1455345_at | 18295 | -1.413 | -0.1553 | No | ||
| 97 | MYH10 | 1441057_at 1452740_at | 18550 | -1.535 | -0.1598 | No | ||
| 98 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 18680 | -1.587 | -0.1583 | No | ||
| 99 | FANCG | 1420960_at | 18683 | -1.587 | -0.1509 | No | ||
| 100 | USP37 | 1434242_at 1438212_at | 18838 | -1.688 | -0.1500 | No | ||
| 101 | CASP8AP2 | 1449217_at | 18948 | -1.755 | -0.1468 | No | ||
| 102 | UGCGL1 | 1432318_at 1448009_at 1455839_at | 19081 | -1.848 | -0.1442 | No | ||
| 103 | ACBD6 | 1452601_a_at 1453199_at | 19094 | -1.855 | -0.1360 | No | ||
| 104 | ZCCHC8 | 1423755_at 1441639_at | 19661 | -2.248 | -0.1514 | No | ||
| 105 | YBX2 | 1420762_a_at | 19728 | -2.310 | -0.1436 | No | ||
| 106 | IL4I1 | 1419192_at | 19954 | -2.535 | -0.1420 | No | ||
| 107 | ZBTB4 | 1429722_at 1460488_at | 20319 | -2.930 | -0.1450 | No | ||
| 108 | MXD3 | 1422970_at 1438847_at 1448878_at | 20338 | -2.963 | -0.1319 | No | ||
| 109 | ALDH6A1 | 1448104_at | 20363 | -2.992 | -0.1190 | No | ||
| 110 | STK35 | 1431107_at 1439802_at | 20364 | -2.993 | -0.1049 | No | ||
| 111 | MAZ | 1427099_at | 20396 | -3.020 | -0.0922 | No | ||
| 112 | PHC1 | 1423212_at | 20522 | -3.189 | -0.0829 | No | ||
| 113 | TRIM39 | 1418851_at | 21110 | -4.153 | -0.0904 | No | ||
| 114 | H2AFZ | 1441370_at | 21157 | -4.282 | -0.0724 | No | ||
| 115 | USP52 | 1426700_a_at | 21560 | -5.702 | -0.0640 | No | ||
| 116 | DCK | 1428838_a_at 1439012_a_at 1449176_a_at | 21820 | -8.499 | -0.0360 | No | ||
| 117 | ID3 | 1416630_at | 21834 | -8.786 | 0.0046 | No |

