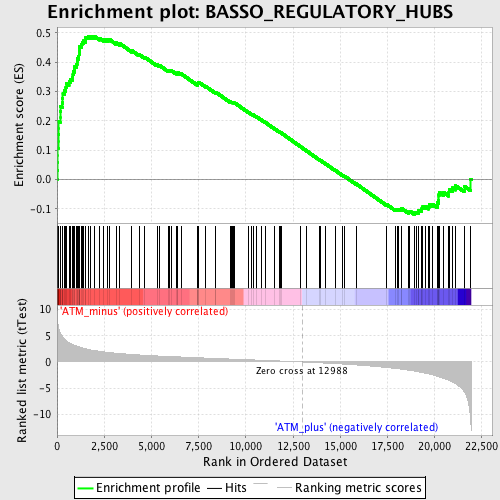
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | BASSO_REGULATORY_HUBS |
| Enrichment Score (ES) | 0.48891744 |
| Normalized Enrichment Score (NES) | 2.0347638 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0054870592 |
| FWER p-Value | 0.043 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TCP1 | 1442317_at 1448122_at | 7 | 9.127 | 0.0309 | Yes | ||
| 2 | AFG3L2 | 1427206_at 1427207_s_at 1454003_at | 34 | 7.841 | 0.0565 | Yes | ||
| 3 | PSMD1 | 1419799_at 1429291_at | 41 | 7.327 | 0.0813 | Yes | ||
| 4 | TOMM40 | 1426118_a_at 1459680_at | 44 | 7.295 | 0.1062 | Yes | ||
| 5 | HDDC2 | 1428823_at | 54 | 7.099 | 0.1301 | Yes | ||
| 6 | CYCS | 1422484_at 1445484_at | 57 | 6.950 | 0.1538 | Yes | ||
| 7 | SLC7A5 | 1418326_at | 80 | 6.452 | 0.1748 | Yes | ||
| 8 | HPRT1 | 1448736_a_at | 85 | 6.404 | 0.1966 | Yes | ||
| 9 | XPOT | 1428772_at 1428949_at 1441681_at 1441682_s_at | 159 | 5.571 | 0.2123 | Yes | ||
| 10 | TRIP13 | 1429294_at 1429295_s_at 1446760_at | 163 | 5.509 | 0.2310 | Yes | ||
| 11 | CTPS | 1416563_at | 178 | 5.392 | 0.2488 | Yes | ||
| 12 | GARS | 1423784_at | 274 | 4.866 | 0.2611 | Yes | ||
| 13 | MRPL12 | 1452048_at | 298 | 4.780 | 0.2764 | Yes | ||
| 14 | PRG1 | 1417426_at | 310 | 4.713 | 0.2920 | Yes | ||
| 15 | JTV1 | 1424151_at 1451262_a_at | 383 | 4.421 | 0.3038 | Yes | ||
| 16 | D15WSU75E | 1457181_at 1460278_a_at 1460689_at | 465 | 4.130 | 0.3142 | Yes | ||
| 17 | TIMM17A | 1426256_at | 493 | 4.032 | 0.3268 | Yes | ||
| 18 | MAD2L1 | 1422460_at | 631 | 3.687 | 0.3331 | Yes | ||
| 19 | PRDX3 | 1416292_at | 711 | 3.539 | 0.3416 | Yes | ||
| 20 | PSMA3 | 1448442_a_at | 798 | 3.372 | 0.3492 | Yes | ||
| 21 | BLMH | 1438961_s_at 1439325_at 1452101_at | 839 | 3.299 | 0.3587 | Yes | ||
| 22 | DBF4 | 1418334_at 1430545_at | 860 | 3.265 | 0.3689 | Yes | ||
| 23 | PAICS | 1423564_a_at 1423565_at 1436298_x_at | 921 | 3.157 | 0.3770 | Yes | ||
| 24 | ARVCF | 1423061_at 1450855_at 1450856_at | 939 | 3.133 | 0.3869 | Yes | ||
| 25 | RCC1 | 1416962_at | 1036 | 3.009 | 0.3928 | Yes | ||
| 26 | CCNA2 | 1417910_at 1417911_at | 1058 | 2.987 | 0.4021 | Yes | ||
| 27 | TEP1 | 1418196_at | 1069 | 2.979 | 0.4118 | Yes | ||
| 28 | RFC4 | 1424321_at 1438161_s_at | 1141 | 2.887 | 0.4184 | Yes | ||
| 29 | HSPE1 | 1450668_s_at | 1159 | 2.872 | 0.4275 | Yes | ||
| 30 | CYC1 | 1416604_at | 1168 | 2.860 | 0.4369 | Yes | ||
| 31 | RUVBL2 | 1422482_at | 1191 | 2.827 | 0.4455 | Yes | ||
| 32 | TPX2 | 1428104_at 1428105_at 1447518_at 1447519_x_at | 1200 | 2.817 | 0.4548 | Yes | ||
| 33 | CSE1L | 1448809_at | 1291 | 2.731 | 0.4600 | Yes | ||
| 34 | PCNA | 1417947_at | 1322 | 2.702 | 0.4679 | Yes | ||
| 35 | LOR | 1420183_at 1448745_s_at | 1415 | 2.604 | 0.4726 | Yes | ||
| 36 | LGALS9 | 1421217_a_at | 1517 | 2.506 | 0.4765 | Yes | ||
| 37 | BUD31 | 1433611_s_at 1439466_s_at | 1519 | 2.506 | 0.4851 | Yes | ||
| 38 | CCNB1 | 1419943_s_at 1419944_at | 1648 | 2.393 | 0.4874 | Yes | ||
| 39 | HNRPAB | 1415914_at 1426114_at 1431349_at 1448144_at 1453849_s_at 1455855_x_at 1459880_at | 1786 | 2.286 | 0.4889 | Yes | ||
| 40 | LSM3 | 1448536_at | 1971 | 2.179 | 0.4879 | No | ||
| 41 | AHSA1 | 1424147_at 1444500_at | 2243 | 2.028 | 0.4825 | No | ||
| 42 | MRPS12 | 1448799_s_at | 2475 | 1.912 | 0.4784 | No | ||
| 43 | SAR1A | 1415696_at 1423720_a_at | 2657 | 1.841 | 0.4764 | No | ||
| 44 | SSRP1 | 1426788_a_at 1426789_s_at 1426790_at | 2750 | 1.805 | 0.4784 | No | ||
| 45 | MRPL3 | 1422463_a_at 1422464_at 1431006_at 1440636_at | 3135 | 1.669 | 0.4665 | No | ||
| 46 | SSBP1 | 1427965_at 1430294_at 1444053_at 1452616_s_at 1452617_at | 3328 | 1.614 | 0.4632 | No | ||
| 47 | ASNS | 1433966_x_at 1451095_at | 3957 | 1.431 | 0.4393 | No | ||
| 48 | NDRG1 | 1423413_at 1450976_at 1456174_x_at | 4343 | 1.341 | 0.4262 | No | ||
| 49 | LGALS1 | 1419573_a_at 1455439_a_at | 4643 | 1.267 | 0.4169 | No | ||
| 50 | FEN1 | 1421731_a_at 1436454_x_at | 5289 | 1.144 | 0.3912 | No | ||
| 51 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 5441 | 1.119 | 0.3881 | No | ||
| 52 | S100A4 | 1424542_at | 5878 | 1.044 | 0.3717 | No | ||
| 53 | POLR2H | 1424473_at | 5948 | 1.035 | 0.3721 | No | ||
| 54 | ORC5L | 1415830_at 1441601_at | 6040 | 1.020 | 0.3714 | No | ||
| 55 | LAPTM5 | 1426025_s_at 1436905_x_at 1447742_at 1459841_x_at | 6303 | 0.982 | 0.3627 | No | ||
| 56 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 6384 | 0.968 | 0.3624 | No | ||
| 57 | PSMB7 | 1416240_at | 6396 | 0.967 | 0.3652 | No | ||
| 58 | POLD2 | 1448277_at | 6571 | 0.942 | 0.3604 | No | ||
| 59 | NME1 | 1424110_a_at 1435277_x_at | 7443 | 0.804 | 0.3232 | No | ||
| 60 | RPS26 | 1415876_a_at | 7447 | 0.803 | 0.3259 | No | ||
| 61 | PRDX4 | 1416166_a_at 1416167_at 1431269_at | 7453 | 0.803 | 0.3284 | No | ||
| 62 | POLR2F | 1415754_at 1427402_at | 7473 | 0.800 | 0.3302 | No | ||
| 63 | BYSL | 1422767_at 1450742_at | 7503 | 0.795 | 0.3316 | No | ||
| 64 | TUBG1 | 1417144_at | 7867 | 0.744 | 0.3175 | No | ||
| 65 | LITAF | 1416303_at 1416304_at | 8372 | 0.667 | 0.2967 | No | ||
| 66 | PRKCB1 | 1423478_at 1438981_at 1443144_at 1459674_at 1460419_a_at | 8413 | 0.661 | 0.2971 | No | ||
| 67 | MVP | 1448618_at 1456586_x_at | 9164 | 0.552 | 0.2646 | No | ||
| 68 | CTSH | 1418365_at 1443814_x_at | 9226 | 0.543 | 0.2637 | No | ||
| 69 | CASP4 | 1449591_at | 9293 | 0.532 | 0.2625 | No | ||
| 70 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 9315 | 0.529 | 0.2634 | No | ||
| 71 | FAIM3 | 1429889_at | 9412 | 0.517 | 0.2607 | No | ||
| 72 | HHEX | 1423319_at | 10155 | 0.417 | 0.2281 | No | ||
| 73 | UCHL1 | 1448260_at | 10304 | 0.397 | 0.2227 | No | ||
| 74 | SERBP1 | 1418563_at 1418564_s_at 1418565_at 1437280_s_at 1440195_at 1441501_at | 10376 | 0.386 | 0.2208 | No | ||
| 75 | PSMB5 | 1415676_a_at | 10566 | 0.356 | 0.2133 | No | ||
| 76 | TPP1 | 1434768_at 1448313_at | 10807 | 0.326 | 0.2034 | No | ||
| 77 | EPOR | 1423344_at | 11038 | 0.295 | 0.1939 | No | ||
| 78 | BHLHB2 | 1418025_at | 11517 | 0.223 | 0.1728 | No | ||
| 79 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 11802 | 0.182 | 0.1604 | No | ||
| 80 | SHFM1 | 1418574_a_at 1418575_at | 11832 | 0.177 | 0.1596 | No | ||
| 81 | ITPR2 | 1421678_at 1424833_at 1424834_s_at 1427287_s_at 1427693_at 1444418_at 1444551_at 1447328_at | 11866 | 0.171 | 0.1587 | No | ||
| 82 | RBM5 | 1438069_a_at 1441319_at 1444618_at 1444971_at 1452186_at 1452187_at 1456262_at | 12872 | 0.022 | 0.1127 | No | ||
| 83 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 13197 | -0.035 | 0.0980 | No | ||
| 84 | SLC4A3 | 1418485_at 1453687_at | 13905 | -0.161 | 0.0661 | No | ||
| 85 | AHNAK | 1428057_a_at 1428058_at 1429866_at 1452217_at | 13948 | -0.168 | 0.0648 | No | ||
| 86 | ZMYND11 | 1426531_at 1426532_at 1436153_a_at 1444461_at 1446182_at | 14200 | -0.214 | 0.0540 | No | ||
| 87 | ZBTB20 | 1422064_a_at 1437065_at 1437066_at 1437598_at 1438443_at 1439128_at 1439278_at 1440565_at 1443471_at 1451577_at | 14750 | -0.318 | 0.0299 | No | ||
| 88 | CRIP1 | 1416326_at | 15111 | -0.389 | 0.0147 | No | ||
| 89 | UBE2S | 1416726_s_at 1430962_at | 15210 | -0.408 | 0.0116 | No | ||
| 90 | PSMD2 | 1415831_at | 15833 | -0.563 | -0.0150 | No | ||
| 91 | CYLD | 1429617_at 1429618_at 1445213_at | 17466 | -1.050 | -0.0862 | No | ||
| 92 | WDR18 | 1433635_at 1443758_at 1454695_at 1460622_x_at | 17930 | -1.247 | -0.1031 | No | ||
| 93 | CTNNA1 | 1437275_at 1437807_x_at 1443662_at 1448149_at | 18012 | -1.280 | -0.1025 | No | ||
| 94 | SLC39A14 | 1425649_at 1427035_at 1438490_at 1457770_at | 18104 | -1.313 | -0.1022 | No | ||
| 95 | TCEB1 | 1427914_a_at 1427915_s_at 1436226_at 1452593_a_at | 18220 | -1.374 | -0.1027 | No | ||
| 96 | DDB2 | 1425706_a_at 1436680_s_at | 18237 | -1.379 | -0.0987 | No | ||
| 97 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18631 | -1.564 | -0.1114 | No | ||
| 98 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 18680 | -1.587 | -0.1082 | No | ||
| 99 | BIN1 | 1423618_at 1425532_a_at 1444120_at | 18931 | -1.744 | -0.1137 | No | ||
| 100 | EIF2S1 | 1440272_at 1452662_a_at | 19026 | -1.817 | -0.1118 | No | ||
| 101 | SP100 | 1434457_at 1447213_at 1451821_a_at | 19131 | -1.887 | -0.1101 | No | ||
| 102 | USP4 | 1442319_at 1450892_a_at | 19151 | -1.903 | -0.1044 | No | ||
| 103 | P2RY10 | 1452815_at | 19285 | -1.998 | -0.1037 | No | ||
| 104 | TNFRSF1B | 1418099_at 1448951_at | 19320 | -2.022 | -0.0983 | No | ||
| 105 | NISCH | 1430151_at 1433757_a_at 1433758_at 1451338_at 1452156_a_at 1457767_at | 19372 | -2.056 | -0.0936 | No | ||
| 106 | EBNA1BP2 | 1428315_at 1437512_x_at 1439898_at 1457668_x_at | 19500 | -2.128 | -0.0922 | No | ||
| 107 | MDFIC | 1427040_at | 19694 | -2.276 | -0.0932 | No | ||
| 108 | UCK2 | 1417137_at 1426909_at 1439740_s_at 1439741_x_at 1441858_at 1448604_at 1457980_x_at | 19716 | -2.300 | -0.0863 | No | ||
| 109 | PLCB2 | 1452481_at | 19874 | -2.451 | -0.0851 | No | ||
| 110 | EEF1E1 | 1444757_at 1449044_at | 20135 | -2.731 | -0.0877 | No | ||
| 111 | MGEA5 | 1422900_at 1422901_at 1422902_s_at 1433654_at 1439081_at | 20154 | -2.753 | -0.0791 | No | ||
| 112 | ITM2A | 1423608_at 1451047_at | 20202 | -2.799 | -0.0717 | No | ||
| 113 | EMG1 | 1416393_at | 20217 | -2.820 | -0.0627 | No | ||
| 114 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 20218 | -2.820 | -0.0531 | No | ||
| 115 | ETHE1 | 1417203_at | 20230 | -2.833 | -0.0439 | No | ||
| 116 | UNC84B | 1433832_at 1439238_at | 20478 | -3.124 | -0.0445 | No | ||
| 117 | PES1 | 1416172_at 1416173_at | 20747 | -3.513 | -0.0448 | No | ||
| 118 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 20754 | -3.528 | -0.0330 | No | ||
| 119 | GABARAP | 1416937_at | 20951 | -3.828 | -0.0289 | No | ||
| 120 | ZYX | 1417240_at 1438552_x_at | 21111 | -4.159 | -0.0219 | No | ||
| 121 | IL27RA | 1449508_at | 21556 | -5.694 | -0.0228 | No | ||
| 122 | ANXA6 | 1415818_at 1429246_a_at 1429247_at 1442288_at | 21906 | -11.714 | 0.0013 | No |

