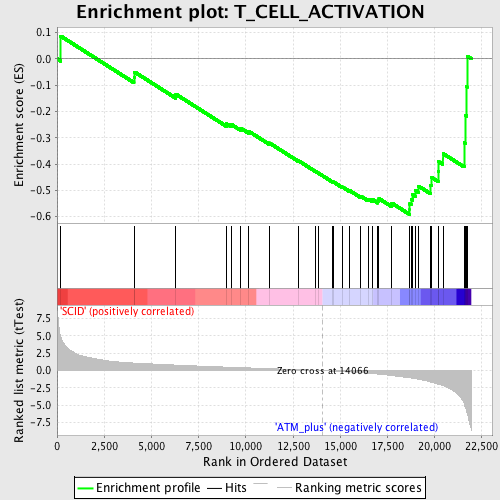
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | T_CELL_ACTIVATION |
| Enrichment Score (ES) | -0.5915828 |
| Normalized Enrichment Score (NES) | -1.80182 |
| Nominal p-value | 0.0019723866 |
| FDR q-value | 0.18521053 |
| FWER p-Value | 0.738 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTPRC | 1422124_a_at 1440165_at | 195 | 4.864 | 0.0860 | No | ||
| 2 | IL4 | 1449864_at | 4073 | 1.082 | -0.0699 | No | ||
| 3 | CLEC7A | 1420699_at | 4107 | 1.076 | -0.0504 | No | ||
| 4 | CD276 | 1417599_at | 6289 | 0.794 | -0.1345 | No | ||
| 5 | ICOSLG | 1419212_at 1426087_at | 8965 | 0.503 | -0.2468 | No | ||
| 6 | IL21 | 1450334_at | 9226 | 0.478 | -0.2494 | No | ||
| 7 | CD3E | 1422105_at 1445748_at | 9711 | 0.429 | -0.2631 | No | ||
| 8 | IL7 | 1422080_at 1436861_at | 10155 | 0.388 | -0.2758 | No | ||
| 9 | NCK2 | 1416796_at 1416797_at 1458432_at | 11246 | 0.287 | -0.3200 | No | ||
| 10 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 12793 | 0.138 | -0.3879 | No | ||
| 11 | IL18 | 1417932_at | 13700 | 0.039 | -0.4285 | No | ||
| 12 | CD47 | 1419554_at 1428187_at 1430997_at 1449507_a_at 1458634_at 1459908_at | 13844 | 0.023 | -0.4345 | No | ||
| 13 | ELF4 | 1421337_at 1445778_at | 14591 | -0.064 | -0.4674 | No | ||
| 14 | SART1 | 1448658_at | 14634 | -0.070 | -0.4679 | No | ||
| 15 | SOCS5 | 1423349_at 1423350_at 1441640_at 1442890_at | 15112 | -0.132 | -0.4871 | No | ||
| 16 | FOXP3 | 1420765_a_at 1457082_at | 15507 | -0.190 | -0.5014 | No | ||
| 17 | SFTPD | 1420378_at | 16076 | -0.286 | -0.5217 | No | ||
| 18 | SPINK5 | 1430567_at | 16476 | -0.364 | -0.5328 | No | ||
| 19 | JAG2 | 1426430_at 1426431_at | 16700 | -0.421 | -0.5348 | No | ||
| 20 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 16992 | -0.494 | -0.5385 | No | ||
| 21 | LAT | 1460651_at | 17045 | -0.505 | -0.5310 | No | ||
| 22 | NHEJ1 | 1425651_at 1429098_s_at | 17727 | -0.707 | -0.5483 | No | ||
| 23 | IL12B | 1419530_at 1449497_at | 18676 | -1.031 | -0.5715 | Yes | ||
| 24 | CRTAM | 1449903_at | 18681 | -1.032 | -0.5515 | Yes | ||
| 25 | SLA2 | 1437504_at 1451492_at | 18752 | -1.059 | -0.5341 | Yes | ||
| 26 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 18832 | -1.093 | -0.5164 | Yes | ||
| 27 | EBI3 | 1449222_at | 18973 | -1.159 | -0.5001 | Yes | ||
| 28 | GLMN | 1418839_at | 19143 | -1.237 | -0.4837 | Yes | ||
| 29 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 19770 | -1.625 | -0.4806 | Yes | ||
| 30 | CD4 | 1419696_at 1427779_a_at | 19831 | -1.668 | -0.4508 | Yes | ||
| 31 | CD28 | 1417597_at 1437025_at 1443703_at | 20183 | -1.956 | -0.4286 | Yes | ||
| 32 | LAX1 | 1438687_at | 20206 | -1.973 | -0.3911 | Yes | ||
| 33 | THY1 | 1423135_at | 20438 | -2.139 | -0.3600 | Yes | ||
| 34 | CD3D | 1422828_at | 21560 | -4.763 | -0.3182 | Yes | ||
| 35 | CD7 | 1419711_at | 21644 | -5.423 | -0.2162 | Yes | ||
| 36 | SIT1 | 1418751_at | 21687 | -5.733 | -0.1063 | Yes | ||
| 37 | CD2 | 1418770_at | 21715 | -6.027 | 0.0101 | Yes |

